-
Notifications
You must be signed in to change notification settings - Fork 4
/
Copy pathauthorisation.Rmd
98 lines (71 loc) · 3.66 KB
/
authorisation.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
---
title: "Authorising yourself for neuPrint"
output: rmarkdown::html_vignette
vignette: >
%\VignetteIndexEntry{Authorising yourself for neuPrint}
%\VignetteEngine{knitr::rmarkdown}
%\VignetteEncoding{UTF-8}
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>"
)
```
## Installation
```{r install, eval = FALSE}
# install
if (!require("devtools")) install.packages("devtools")
devtools::install_github("natverse/neuprintr")
# use
library(neuprintr)
```
## Authentication
In order to use *neuprintr* you will need to be able to login to a neuPrint server and be able to access it underlying Neo4j database.
You may need an authenticated accounted, or you may be able to register your `@gmail` address without an authentication process.
Navigate to a neuPrint website, e.g. https://neuprint.janelia.org, and hit 'login'. Sign in using an `@gmail` account.
If you have authentication/the server is public, you will now be able to see your access token by going to 'Account':
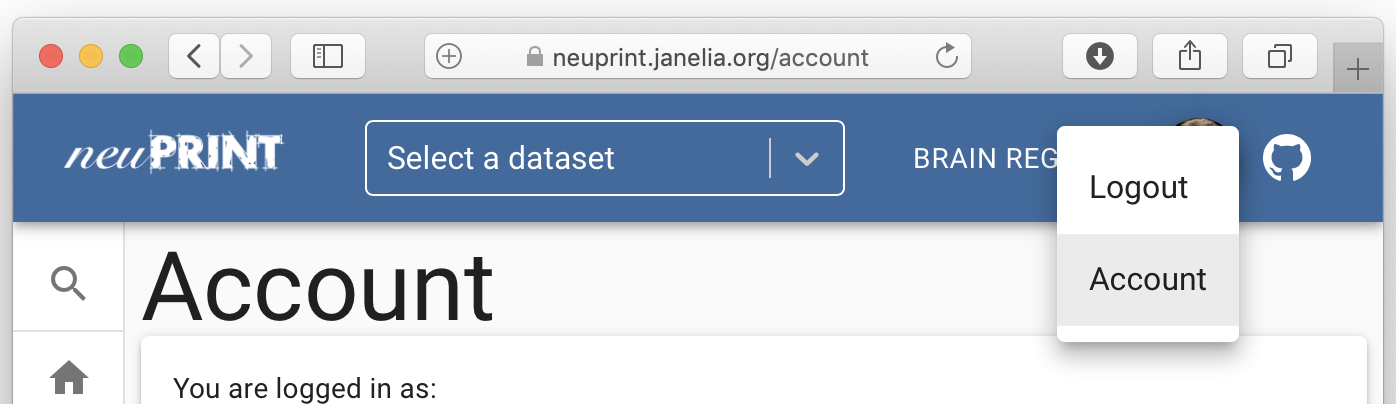
To make life easier, you can then edit your `.Renviron` file to contain information about the neuPrint server you want to speak with, your token and the dataset hosted by that server, that you want to read. A convenient way to do this is to do
```{r, eval=FALSE}
usethis::edit_r_environ()
```
and then edit the file that pops up, adding a section like
```{r, eval=FALSE}
neuprint_server="https://neuprint.janelia.org"
# nb this token is a dummy
neuprint_token="asBatEsiOIJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJlbWFpbCI6ImIsImxldmVsIjoicmVhZHdyaXRlIiwiaW1hZ2UtdXJsIjoiaHR0cHM7Ly9saDQuZ29vZ2xldXNlcmNvbnRlbnQuY29tLy1QeFVrTFZtbHdmcy9BQUFBQUFBQUFBDD9BQUFBQUFBQUFBQS9BQ0hpM3JleFZMeEI4Nl9FT1asb0dyMnV0QjJBcFJSZlI6MTczMjc1MjU2HH0.jhh1nMDBPl5A1HYKcszXM518NZeAhZG9jKy3hzVOWEU"
```
Make sure you have a blank line at the end of your `.Renviron` file. Note that
you can optionally specify a default dataset:
```{r, eval=FALSE}
neuprint_dataset = "hemibrain:v1.0"
```
if your neuPrint server has more than one dataset. For further information
about neuprintr login, see the help for `neuprint_login()`.
Finally you can also login on the command line once per session, like so:
```{r login2, eval = FALSE}
conn = neuprint_login(server= "https://neuprint.janelia.org/",
token= "asBatEsiOIJIUzI1NiIsInR5cCI6IkpXVCJ9.eyJlbWFpbCI6ImIsImxldmVsIjoicmVhZHdyaXRlIiwiaW1hZ2UtdXJsIjoiaHR0cHM7Ly9saDQuZ29vZ2xldXNlcmNvbnRlbnQuY29tLy1QeFVrTFZtbHdmcy9BQUFBQUFBQUFBDD9BQUFBQUFBQUFBQS9BQ0hpM3JleFZMeEI4Nl9FT1asb0dyMnV0QjJBcFJSZlI6MTczMjc1MjU2HH0.jhh1nMDBPl5A1HYKcszXM518NZeAhZG9jKy3hzVOWEU")
```
This is also the approach that you would take if you were working with more than
two neuPrint servers.
## Example
Now we can have a look at what is available
```{r example, eval = FALSE}
# What data sets are available?
neuprint_datasets()
# What's the underlying database
neuprint_database()
# What are the regions of interrst in your default datasest (specified in R.environ, see ?neuprint_login)
neuprint_ROIs()
```
Use the client to request data from neuprint. The `neuprint_fetch_custom` method will run an arbitrary cypher query against the database. For information about the neuprint data model, see the neuprint explorer web help: https://neuprint.janelia.org/help.
Some cyphers and other API endpoints have been explored by this package. Have a look a the functions, for example, that give you neuron skeletons, synapse locations, connectivity matrices, etc.
```{r example2, eval = FALSE}
?neuprint_search
?neuprint_get_adjacency_matrix
?neuprint_ROI_connectivity
?neuprint_get_synapses
?neuprint_read_neurons
```