1. Load data
We assume you have already read the Introduction and installed wham and its dependencies. If not, you should be able to just run devtools::install_github("timjmiller/wham", dependencies=TRUE).
Open R and load the wham package:
+For a clean, runnable
-.Rscript, look atex1_basics.Rin theexample_scriptsfolder of thewhampackage install:++#> [1] "/tmp/RtmpS2yNsx/temp_libpath5e76603d0384/wham/example_scripts"wham.dir <- find.package("wham") file.path(wham.dir, "example_scripts") -#> [1] "/tmp/RtmpxsYcd5/temp_libpath4c62c90d065/wham/example_scripts"You can run this entire example script with (2.0 min runtime):
-+-write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory source(file.path(wham.dir, "example_scripts", "ex1_basics.R"))Let’s create a directory for this analysis:
-+Let's create a directory for this analysis:
+# choose a location to save output write.dir <- "/path/to/save/output" # modify dir.create(write.dir) setwd(write.dir)WHAM was built by modifying the ADMB-based ASAP model code (Legault and Restrepo 1999), and is designed to take an ASAP3 .dat file as input. We generally assume in
whamthat you have an existing ASAP3 .dat file. If you are not familiar with ASAP3 input files, see the ASAP documentation. For this vignette, an example ASAP3 input file is provided.Copy
-ex1_SNEMAYT.datto our analysis directory:+wham.dir <- find.package("wham") file.copy(from=file.path(wham.dir,"extdata","ex1_SNEMAYT.dat"), to=write.dir, overwrite=FALSE)Confirm you are in the working directory and it has the
-ex1_SNEMAYT.datfile:+list.files() #> [1] "CPI.csv" "ex1_SNEMAYT.dat" "ex1_test_results.rds" #> [4] "ex2_SNEMAYT.dat" "ex2_test_results.rds" "ex4_test_results.rds" #> [7] "ex5_summerflounder.dat" "ex5_test_results.rds" "ex6_test_results.rds" #> [10] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R:
-+asap3 <- read_asap3_dat("ex1_SNEMAYT.dat")@@ -193,7 +193,7 @@
recruitment deviations: independent random effects ( NAA_re = list(sigma="rec", cor="iid"))selectivity: age-specific (fix sel=1 for age 5 in fishery, age 4 in index1, and age 2 in index2) -+input1 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder", selectivity=list(model=rep("age-specific",3), re=rep("none",3), @@ -212,11 +212,11 @@
3. Fit model and check for convergence
-+m1 <- fit_wham(input1, do.osa = F) # turn off OSA residuals to save time in exBy default,
-fit_wham()uses 3 extra Newton steps to reduce the absolute value of the gradient (n.newton = 3) and estimates standard errors for derived parameters (do.sdrep = TRUE).fit_wham()also does a retrospective analysis with 7 peels by default (do.retro = TRUE,n.peels = 7). For more details, seefit_wham().We need to check that
-m1converged (m1$opt$convergenceshould be 0, and the maximum absolute value of the gradient vector should be < 1e-06). Convergence issues may indicate that a model is misspecified or overparameterized. To help diagnose these problems,fit_wham()includes ado.checkoption to run Jim Thorson’s useful function TMBhelper::Check_Identifiable().do.check = FALSEby default. To turn on, setdo.check = TRUE. Seefit_wham().+We need to check that
+m1converged (m1$opt$convergenceshould be 0, and the maximum absolute value of the gradient vector should be < 1e-06). Convergence issues may indicate that a model is misspecified or overparameterized. To help diagnose these problems,fit_wham()includes ado.checkoption to run Jim Thorson's useful function TMBhelper::Check_Identifiable().do.check = FALSEby default. To turn on, setdo.check = TRUE. Seefit_wham().#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0 #> Maximum gradient component: 1.20e-07 @@ -227,7 +227,7 @@Fit models
m2-m4The second model,
-m2, is like the first, but changes all the age composition likelihoods from multinomial to logistic normal (treating 0 observations as missing):+input2 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder", selectivity=list(model=rep("age-specific",3), re=rep("none",3), @@ -237,14 +237,14 @@age_comp = "logistic-normal-miss0") m2 <- fit_wham(input2, do.osa = F) # turn off OSA residuals to save time in ex
Check that
-m2converged:+#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0 #> Maximum gradient component: 4.96e-12 #> Max gradient parameter: logit_selpars #> TMB:sdreport() was performed successfully for this modelThe third,
-m3, is a full state-space model where numbers at all ages are random effects (NAA_re$sigma = "rec+1"):+input3 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder", selectivity=list(model=rep("age-specific",3), re=rep("none",3), @@ -253,14 +253,14 @@NAA_re = list(sigma="rec+1", cor="iid")) m3 <- fit_wham(input3, do.osa = F) # turn off OSA residuals to save time in ex
Check that
-m3converged:+#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0 #> Maximum gradient component: 2.67e-06 #> Max gradient parameter: log_F1 #> TMB:sdreport() was performed successfully for this modelThe last,
-m4, is likem3, but again changes all the age composition likelihoods to logistic normal:+input4 <- prepare_wham_input(asap3, recruit_model=2, model_name="Ex 1: SNEMA Yellowtail Flounder", selectivity=list(model=rep("age-specific",3), re=rep("none",3), @@ -270,40 +270,40 @@age_comp = "logistic-normal-miss0") m4 <- fit_wham(input4, do.osa = F) # turn off OSA residuals to save time in ex
Check that
-m4converged:+#> stats:nlminb thinks the model has converged: mod$opt$convergence == 0 #> Maximum gradient component: 2.32e-07 #> Max gradient parameter: log_F1 #> TMB:sdreport() was performed successfully for this modelStore all models together in one (named) list:
-+mods <- list(m1=m1, m2=m2, m3=m3, m4=m4)Since the retrospective analyses take a few minutes to run, you may want to save the output for later use:
-+save("mods", file="ex1_models.RData")4. Compare models
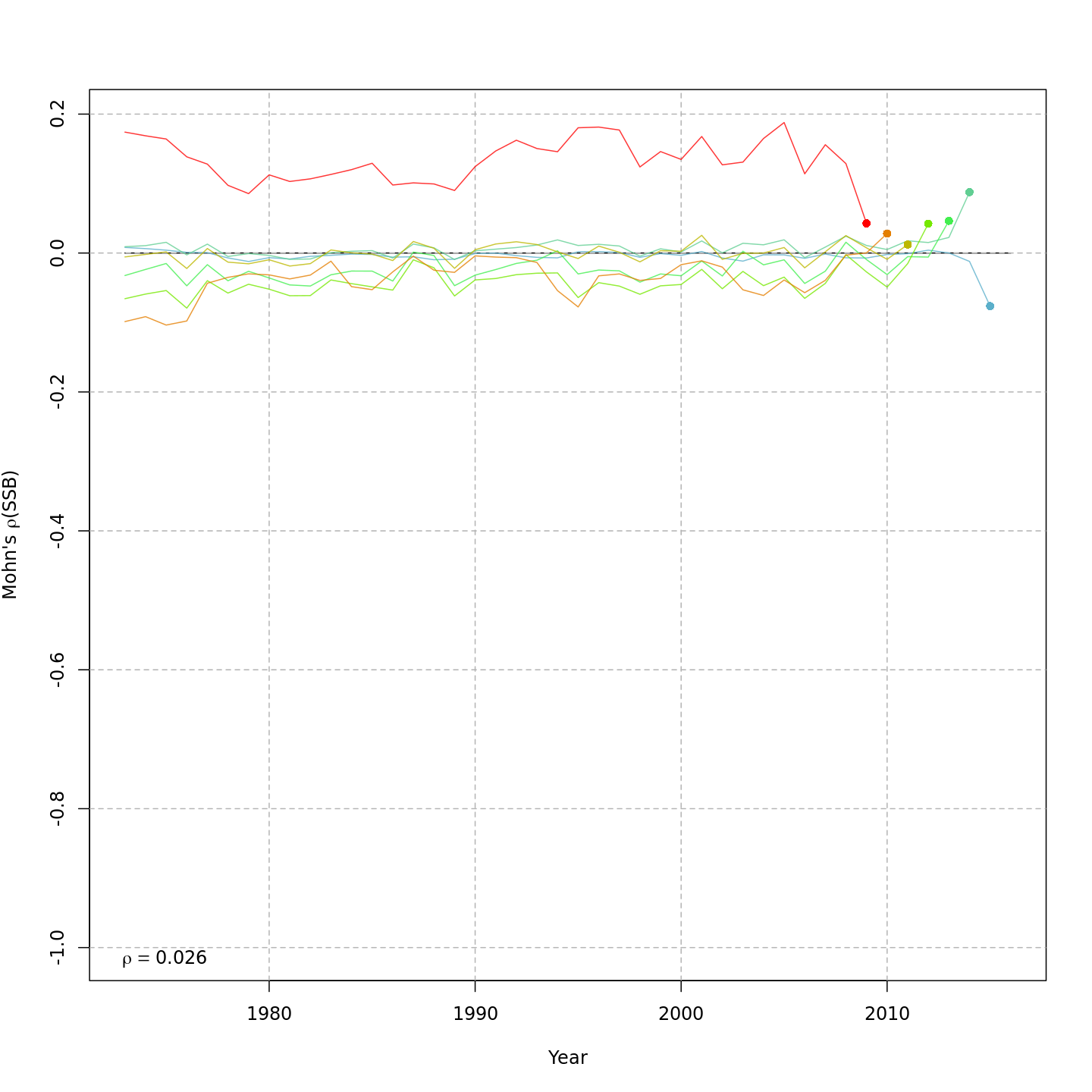
-We can compare models using AIC and Mohn’s rho:
-+We can compare models using AIC and Mohn's rho:
+res <- compare_wham_models(mods, fname="model_comparison", sort=TRUE)-#> dAIC AIC rho_R rho_SSB rho_Fbar #> m4 0.0 -1475.5 0.2918 0.0240 -0.0247 #> m2 302.8 -1172.7 3.1666 -0.0708 0.1510 #> m3 5575.9 4100.4 0.1124 0.0056 -0.0056 #> m1 6322.0 4846.5 0.8207 0.1905 -0.1748+res$best#> [1] "m4"By default,
compare_wham_models()sorts the model comparison table with lowest (best) AIC at the top, and saves it asmodel_comparison.csv. However, in this example the models with alternative likelihoods for the age composition observations are not comparable due to differences in how the observations are defined. Still, the models that treat the age composition observations in the same way can be compared to evaluate whether stochasticity in abundances at age provides better performance, i.e.m1vs.m3(both multinomial) andm2vs.m4(both logistic normal).@@ -311,15 +311,15 @@Project the best model
-Let’s do projections for the best model,
-m4, using the default settings (seeproject_wham()):+Let's do projections for the best model,
+m4, using the default settings (seeproject_wham()):m4_proj <- project_wham(model=m4)
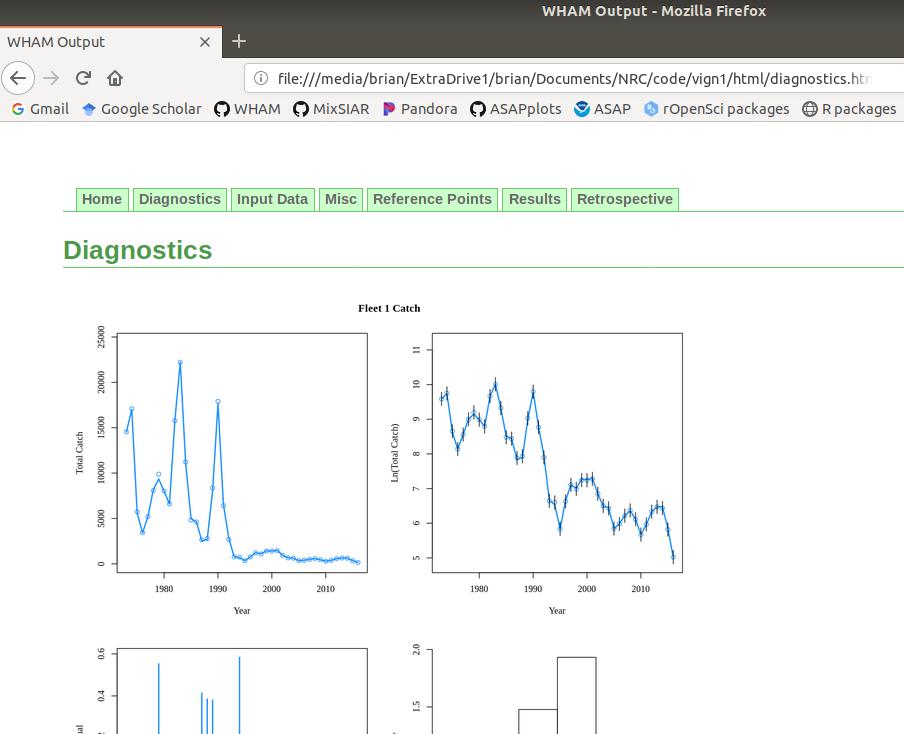
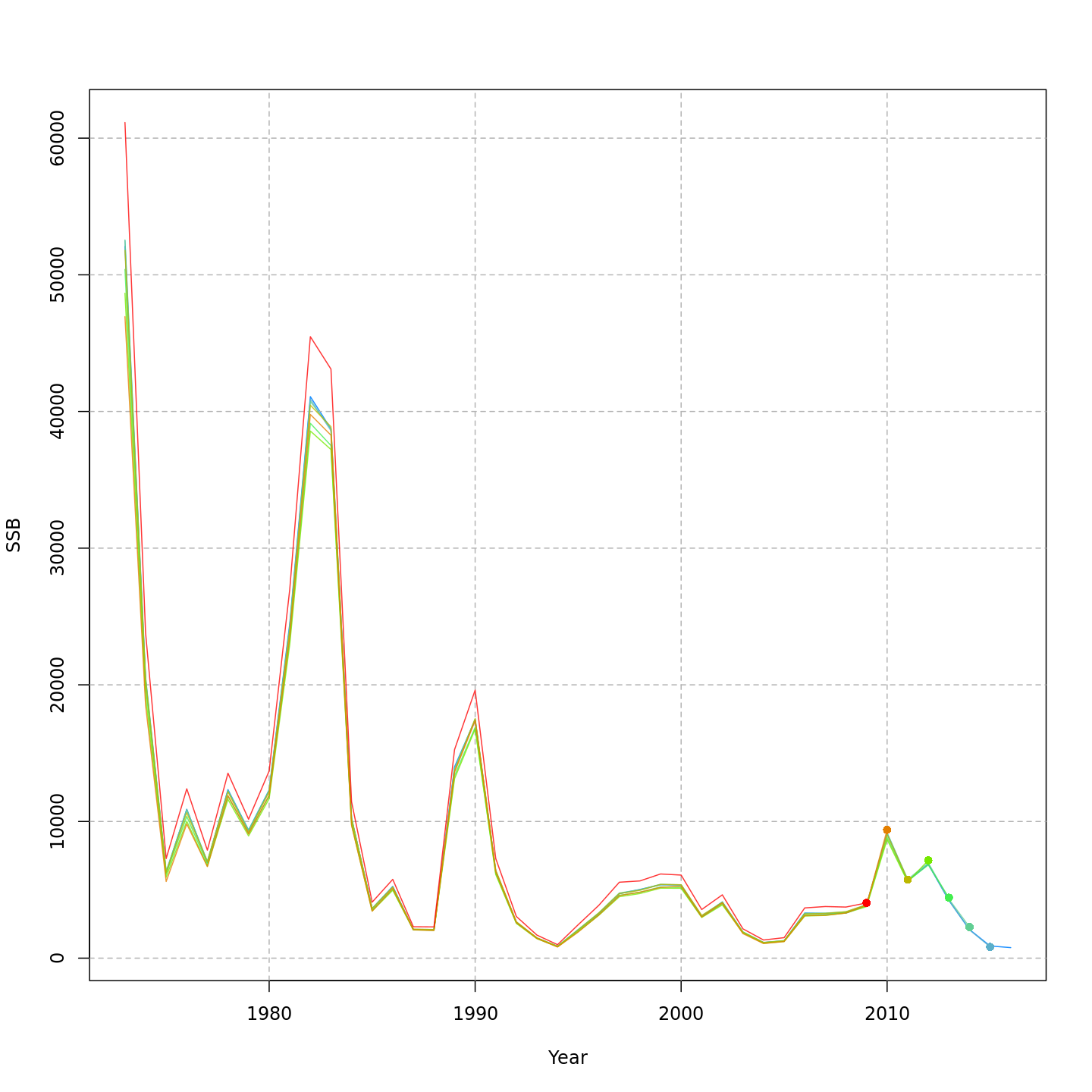
5. Plot input data, diagnostics, and results
There are 3 options for plotting WHAM output. The default (
-out.type='html') creates and opens an HTML file with plots organized into tabs (code modified fromr4ss::SS_html()):+plot_wham_output(mod=m4_proj, out.type='html')Example HTML file created by
plot_wham_output(mod=m4, out.type='html').Setting
-out.type='pdf'saves the plots organized into 6.htmlfile (Diagnostics, Input Data, Results, Reference Points, Retrospective, and Misc). Settingout.type='png'saves the plots as.pngfiles organized into 6 subdirectories.+-plot_wham_output(mod=m4_proj, out.type='png')Many plots are generated—here we display some examples:
+Many plots are generated---here we display some examples:
Diagnostics
@@ -347,7 +347,7 @@
diff --git a/docs/articles/ex2_CPI_recruitment.html b/docs/articles/ex2_CPI_recruitment.html index db947c10..b1f0d33c 100644 --- a/docs/articles/ex2_CPI_recruitment.html +++ b/docs/articles/ex2_CPI_recruitment.html @@ -138,49 +138,49 @@
Ex 2: Recruitment linked to an environmental covariate (Cold P
environmental covariate (Cold Pool Index, CPI) process models (random walk, AR1), and
- how the CPI affects recruitment (controlling or limiting)
As in example 1, we check that each model converges (
+check_convergence()), plot diagnostics, results, and reference points (plot_wham_output()), and compare models using AIC and Mohn’s rho (compare_wham_models()).As in example 1, we check that each model converges (
check_convergence()), plot diagnostics, results, and reference points (plot_wham_output()), and compare models using AIC and Mohn's rho (compare_wham_models()).1. Prepare
whamOpen R and load the
-whampackage:+For a clean, runnable
-.Rscript, look atex2_CPI_recruitment.Rin theexample_scriptsfolder of thewhampackage install:+wham.dir <- find.package("wham") file.path(wham.dir, "example_scripts")You can run this entire example script with:
-+-write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory source(file.path(wham.dir, "example_scripts", "ex2_CPI_recruitment.R"))Let’s create a directory for this analysis:
-+Let's create a directory for this analysis:
+# choose a location to save output, otherwise will be saved in working directory write.dir <- "choose/where/to/save/output" dir.create(write.dir) setwd(write.dir)WHAM was built by modifying the ADMB-based ASAP model code (Legault and Restrepo 1999), and is designed to take an ASAP3 .dat file as input. We generally assume in
whamthat you have an existing ASAP3 .dat file. If you are not familiar with ASAP3 input files, see the ASAP documentation. For this vignette, an example ASAP3 input file is provided,ex2_SNEMAYT.dat. We will also need a data file with an environmental covariate, the Cold Pool Index,CPI.csv.Copy
-ex2_SNEMAYT.datandCPI.csvto our analysis directory:+wham.dir <- find.package("wham") file.copy(from=file.path(wham.dir,"extdata","ex2_SNEMAYT.dat"), to=write.dir, overwrite=FALSE) file.copy(from=file.path(wham.dir,"extdata","CPI.csv"), to=write.dir, overwrite=FALSE)Confirm you are in the correct directory and it has the required data files:
-+list.files() #> [1] "CPI.csv" "ex1_SNEMAYT.dat" "ex1_test_results.rds" #> [4] "ex2_SNEMAYT.dat" "ex2_test_results.rds" "ex4_test_results.rds" #> [7] "ex5_summerflounder.dat" "ex5_test_results.rds" "ex6_test_results.rds" #> [10] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R and convert to input list for wham:
-+asap3 <- read_asap3_dat("ex2_SNEMAYT.dat")Load the environmental covariate (Cold Pool Index, CPI) data into R:
-+-env.dat <- read.csv("CPI.csv", header=T)We generally abbreviate ‘environmental covariate’ as
-ecovin the code. In this example, theecovdata file has columns for observations (CPI), standard error (CPI_sigma), and year (Year). Observations and year are always required. Standard error can be treated as fixed/data with yearly values (as here) or one overall value shared among years. It can also be estimated as a parameter(s), likewise either as yearly values or one overall value.+We generally abbreviate 'environmental covariate' as
+ecovin the code. In this example, theecovdata file has columns for observations (CPI), standard error (CPI_sigma), and year (Year). Observations and year are always required. Standard error can be treated as fixed/data with yearly values (as here) or one overall value shared among years. It can also be estimated as a parameter(s), likewise either as yearly values or one overall value.head(env.dat) #> Year CPI CPI_sigma #> 1 1973 0.5988 0.2838 @@ -194,7 +194,7 @@
2. Specify models
Now we specify how the 7 models treat recruitment, the CPI process, and how the CPI affects recruitment:
-+df.mods <- data.frame(Recruitment = c(2,2,3,3,3,3,4), ecov_process = c(rep("rw",4),rep("ar1",3)), ecov_how = c(0,1,0,2,2,1,1), stringsAsFactors=FALSE) @@ -202,7 +202,7 @@df.mods$Model <- paste0("m",1:n.mods) df.mods <- dplyr::select(df.mods, Model, tidyselect::everything()) # moves Model to first col
Look at the model table
-+df.mods #> Model Recruitment ecov_process ecov_how #> 1 m1 2 rw 0 @@ -220,7 +220,7 @@
Ricker. The environmental covariate options are fed to
-prepare_wham_input()as a list,ecov:+m=1 # example for first model ecov <- list( label = "CPI", @@ -234,23 +234,23 @@how = df.mods$ecov_how[m]) # 0 = no effect, 1 = controlling, 2 = limiting
There are currently 2 options for the
ecovprocess model (ecov$process_model): 1) random walk ('rw'), and 2) autoregressive ('ar1'). We must next specify where theecovaffects the population; here it is via recruitment (ecov$where = "recruit") as opposed to another process like catchability, mortality, maturity, etc. The options for how theecovaffects recruitment (ecov$how) follow Iles and Beverton (1998) and Xu et al. (2018):-
-- “controlling” (dens-indep mortality),
-- “limiting” (carrying capacity, e.g.
-ecovdetermines amount of suitable habitat),- “lethal” (threshold, i.e. R –> 0 at some
ecovvalue),
+- "controlling" (dens-indep mortality),
+- "limiting" (carrying capacity, e.g.
+ecovdetermines amount of suitable habitat),- "lethal" (threshold, i.e. R --> 0 at some
-ecovvalue),
- “masking” (metabolic/growth,
-ecovdecreases dR/dS), and- “directive” (e.g. behavioral).
+- "masking" (metabolic/growth,
+ecovdecreases dR/dS), and- "directive" (e.g. behavioral).
Finally, we specify the lag at which CPI affects recruitment (
+ecov$lag = 1, i.e. CPI in year t affects recruitment in year t + 1).Finally, we specify the lag at which CPI affects recruitment (
ecov$lag = 1, i.e. CPI in year t affects recruitment in year t + 1).You can set
ecov = NULLto fit the model without environmental covariate data, but note that here we fit theecovdata even for models without anecoveffect on recruitment (m1andm3) so that we can compare them via AIC (need to have the same data in the likelihood). We accomplish this by settingecov$how = 0.Options are described in the
-prepare_wham_input()help page. Not allecov$howoptions are implemented for every recruitment model.+?prepare_wham_input3. Run the models
-+for(m in 1:n.mods){ # set up environmental covariate data and model options ecov <- list( @@ -293,11 +293,11 @@
4. Check for convergence
Collect all models into a list.
-+There is no indication that any of the models failed to converge. In addition, SE estimates are calculable for all models (invertible Hessian,
-TMB::sdreport()succeeds).+vign2_conv <- lapply(mods, function(x) capture.output(check_convergence(x))) for(m in 1:n.mods) cat(paste0("Model ",m,":"), vign2_conv[[m]], "", sep='\n')#> Model 1: @@ -345,12 +345,12 @@
5. Compare models
-Calculate AIC and Mohn’s rho using
-compare_wham_models().+Calculate AIC and Mohn's rho using
+compare_wham_models().df.aic <- compare_wham_models(mods, sort=FALSE)$tab # can't sort yet bc going to make labels prettier df.mods <- cbind(df.mods, df.aic)Print and save the results table.
-m6has the lowest AIC (Bev-Holt recruitment, CPI modeled as AR1, controlling effect of CPI on recruitment).+# make results table prettier rownames(df.mods) <- NULL df.mods$Recruitment <- dplyr::recode(df.mods$Recruitment, `2`='Random', `3`='Bev-Holt', `4`='Ricker') @@ -381,7 +381,7 @@
6. Results
There are 3 options for plotting WHAM output. The default (
-out.type='html') creates and opens an HTML file with plots organized into tabs (code modified fromr4ss::SS_html()):+# save output plots in subfolder for each model for(m in 1:n.mods) plot_wham_output(mod=mods[[m]], dir.main=file.path(getwd(), df.mods$Model[m]), out.type='html')diff --git a/docs/articles/ex3_projections.html b/docs/articles/ex3_projections.html index fe95e4a2..3e2f03e5 100644 --- a/docs/articles/ex3_projections.html +++ b/docs/articles/ex3_projections.html @@ -137,7 +137,7 @@Ex 3: Projecting / forecasting random effects
logistic normal age compositions (
input$data$age_comp_model_fleets = 5andinput$data$age_comp_model_indices = 5)Beverton-Holt recruitment (
recruit_model = 3)- Cold Pool Index (CPI) fit as an AR1 process (
ecov$process_model = "ar1")+ CPI has a “controlling” (density-independent mortality, Iles and Beverton (1998)) effect on recruitment (
ecov$where = "recruit",ecov$how = 1)CPI has a "controlling" (density-independent mortality, Iles and Beverton (1998)) effect on recruitment (
ecov$where = "recruit",ecov$how = 1)In example 3, we demonstrate how to project/forecast WHAM models using the
project_wham()function options for handling@@ -148,39 +148,39 @@
Ex 3: Projecting / forecasting random effects
1. Load data
Open R and load the
-whampackage:+For a clean, runnable
-.Rscript, look atex3_projections.Rin theexample_scriptsfolder of thewhampackage install:+wham.dir <- find.package("wham") file.path(wham.dir, "example_scripts")You can run this entire example script with:
-+-write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory source(file.path(wham.dir, "example_scripts", "ex3_projections.R"))Let’s create a directory for this analysis:
-+Let's create a directory for this analysis:
+-# choose a location to save output, otherwise will be saved in working directory write.dir <- "choose/where/to/save/output" dir.create(write.dir) setwd(write.dir)We need the same data files as in example 2. Let’s copy
-ex2_SNEMAYT.datandCPI.csvto our analysis directory:+We need the same data files as in example 2. Let's copy
+ex2_SNEMAYT.datandCPI.csvto our analysis directory:wham.dir <- find.package("wham") file.copy(from=file.path(wham.dir,"extdata","ex2_SNEMAYT.dat"), to=write.dir, overwrite=FALSE) file.copy(from=file.path(wham.dir,"extdata","CPI.csv"), to=write.dir, overwrite=FALSE)Confirm you are in the correct directory and it has the required data files:
-+list.files() #> [1] "CPI.csv" "ex1_SNEMAYT.dat" "ex1_test_results.rds" #> [4] "ex2_SNEMAYT.dat" "ex2_test_results.rds" "ex4_test_results.rds" #> [7] "ex5_summerflounder.dat" "ex5_test_results.rds" "ex6_test_results.rds" #> [10] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R and convert to input list for wham:
-+asap3 <- read_asap3_dat("ex2_SNEMAYT.dat")Load the environmental covariate (Cold Pool Index, CPI) data into R:
-+env.dat <- read.csv("CPI.csv", header=T)@@ -192,9 +192,9 @@
logistic normal age compositions (
input$data$age_comp_model_fleets = 5andinput$data$age_comp_model_indices = 5)Beverton-Holt recruitment (
recruit_model = 3)- -
Cold Pool Index (CPI) fit as an AR1 process (
ecov$process_model = "ar1")- +
CPI has a “controlling” (density-independent mortality, Iles and Beverton (1998)) effect on recruitment (
ecov$where = "recruit",ecov$how = 1)- -
CPI has a "controlling" (density-independent mortality, Iles and Beverton (1998)) effect on recruitment (
ecov$where = "recruit",ecov$how = 1)+env <- list( label = "CPI", mean = as.matrix(env.dat$CPI), # CPI observations @@ -224,18 +224,18 @@
-
- Fit model without projections and then add projections afterward
+# don't run mod <- fit_wham(input) # default do.proj=FALSE mod_proj <- project_wham(mod)-
- Add projections with initial model fit (
do.proj = TRUE)+# don't run mod_proj <- fit_wham(input, do.proj = TRUE)The two code blocks above are equivalent; when
-do.proj = TRUE,fit_wham()fits the model without projections and then callsproject_wham()to add them. In this example we choose option #1 because we are going to add several different projections to the same model,mod. We will save each projected model in a list,mod_proj.+ @@ -244,7 +244,7 @@
4. Add projections to fit model
Projection options are specifed using the
-proj.optsinput toproject_wham(). The default settings are to project 3 years (n.yrs = 3), use average maturity-, weight-, and natural mortality-at-age from last 5 model years to calculate reference points (avg.yrs), use fishing mortality in the last model year (use.last.F = TRUE), and continue the ecov process model (cont.ecov = TRUE). These options are also described in theproject_wham()help page.+# save projected models in a list mod_proj <- list() @@ -257,13 +257,13 @@# mod_proj[[1]] <- project_wham(mod)
WHAM implements four options for handling the environmental covariate(s) in the projections. Exactly one of these must be specified in
proj.optsifecovis in the model:-
- +
(Default) Continue the ecov process model (e.g. random walk, AR1). Set
cont.ecov = TRUE. WHAM will estimate the ecov process in the projection years (i.e. continue the random walk / AR1 process).(Default) Continue the ecov process model (e.g. random walk, AR1). Set
cont.ecov = TRUE. WHAM will estimate the ecov process in the projection years (i.e. continue the random walk / AR1 process).Use last year ecov. Set
use.last.ecov = TRUE. WHAM will use ecov value from the terminal year of the population model for projections.Use average ecov. Provide
avg.yrs.ecov, a vector specifying which years to average over the environmental covariate(s) for projections.Specify ecov. Provide
proj.ecov, a matrix of user-specified environmental covariate(s) to use for projections. Dimensions must be the number of projection years (proj.opts$n.yrs) x the number of ecovs (ncols(ecov$mean)).Note that for all options, if the original model fit the ecov in years beyond the population model, WHAM will use these already-fit ecov values for the projections. If the ecov model extended at least
-proj.opts$n.yrsyears beyond the population model, then none of the above need be specified.+# 5 years, use average ecov from 1992-1996 mod_proj[[2]] <- project_wham(mod, proj.opts=list(n.yrs=5, use.last.F=TRUE, use.avg.F=FALSE, use.FXSPR=FALSE, proj.F=NULL, proj.catch=NULL, avg.yrs=NULL, @@ -305,7 +305,7 @@
Specify F. Provide
proj.F, an F vector with length =proj.opts$n.yrs.- -
Specify catch. Provide
proj.catch, a vector of aggregate catch with length =proj.opts$n.yrs. WHAM will calculate F across fleets to apply the specified catch.+# 5 years, specify catch mod_proj[[6]] <- project_wham(mod, proj.opts=list(n.yrs=5, use.last.F=FALSE, use.avg.F=FALSE, use.FXSPR=FALSE, proj.F=NULL, proj.catch=c(10, 2000, 1000, 3000, 20), avg.yrs=NULL, @@ -341,29 +341,29 @@# equivalent # mod_proj[[10]] <- project_wham(mod, proj.opts=list(n.yrs=10, use.avg.F=TRUE, avg.yrs=1992:1996))
Save projected models
-+saveRDS(mod_proj, file="m6_proj.rds")5. Compare projections
Projecting the model differently should not have changed the marginal negative log-likelihood! Confirm that the NLL is the same for all projected models (within
-1e-06).+mod$opt$obj # original model NLL-#> [1] -829.6624 #> attr(,"logarithm") #> [1] TRUE+nll_proj <- sapply(mod_proj, function(x) x$opt$obj) # projected models NLL round(nll_proj - mod$opt$obj, 6) # difference between original and projected models' NLL-#> [1] 0 0 0 0 0 0 0 0 0 0Now let’s plot results from each of the projected models.
-+Now let's plot results from each of the projected models.
+for(m in 1:length(mod_proj)){ plot_wham_output(mod_proj[[m]], dir.main=file.path(getwd(),paste0("proj_",m)), out.type='html') }To more easily compare the same plots for each projection, we can copy the plots into new folders organized by plot type instead of model.
-+plots <- c("Ecov_1","F_byfleet","SSB_at_age","SSB_F_trend","SSB_Rec_time","Kobe_status") dirs <- file.path(getwd(),plots) lapply(as.list(dirs), FUN=dir.create) diff --git a/docs/articles/ex4_selectivity.html b/docs/articles/ex4_selectivity.html index eb3a7836..2c34d34c 100644 --- a/docs/articles/ex4_selectivity.html +++ b/docs/articles/ex4_selectivity.html @@ -153,42 +153,42 @@Ex 4: Selectivity with time- and age-varying random effects 1. Load data
Open R and load the
-whampackage:+For a clean, runnable
-.Rscript, look atex4_selectivity.Rin theexample_scriptsfolder of thewhampackage install:+wham.dir <- find.package("wham") file.path(wham.dir, "example_scripts")You can run this entire example script with:
-+-write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory source(file.path(wham.dir, "example_scripts", "ex4_selectivity.R"))Let’s create a directory for this analysis:
-+Let's create a directory for this analysis:
+-# choose a location to save output, otherwise will be saved in working directory write.dir <- "choose/where/to/save/output" # need to change dir.create(write.dir) setwd(write.dir)We need the same data files as in example 1. Let’s copy
-ex1_SNEMAYT.datto our analysis directory:+We need the same data files as in example 1. Let's copy
+ex1_SNEMAYT.datto our analysis directory:wham.dir <- find.package("wham") file.copy(from=file.path(wham.dir,"extdata","ex1_SNEMAYT.dat"), to=write.dir, overwrite=FALSE)Confirm you are in the correct directory and it has the required data files:
-+list.files() #> [1] "CPI.csv" "ex1_SNEMAYT.dat" "ex1_test_results.rds" #> [4] "ex2_SNEMAYT.dat" "ex2_test_results.rds" "ex4_test_results.rds" #> [7] "ex5_summerflounder.dat" "ex5_test_results.rds" "ex6_test_results.rds" #> [10] "ex7_SNEMAYT.dat" "GSI.csv"Read the ASAP3 .dat file into R and convert to input list for wham:
-+asap3 <- read_asap3_dat("ex1_SNEMAYT.dat")2. Specify selectivity model options
We are going to run 9 models that differ only in the selectivity options:
-+# m1-m5 logistic, m6-m9 age-specific sel_model <- c(rep("logistic",5), rep("age-specific",4)) @@ -226,7 +226,7 @@
3. Setup and run models
The ASAP data file specifies selectivity options (model, initial parameter values, which parameters to fix/estimate). WHAM uses these by default in order to facilitate running ASAP models. To see the currently specified selectivity options in
-asap3:+asap3$dat$sel_block_assign # 1 fleet, all years assigned to block 1 #> [[1]] #> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 @@ -288,7 +288,7 @@
Model - selectivity$modelNo. Parameters +No. Parameters @@ -318,9 +318,9 @@
Mean selectivity parameters can be initialized at different values from the ASAP file with
selectivity$initial_pars. Parameters can be fixed at their initial values by specifyingselectivity$fix_pars. Finally, we specify any time-varying (random effects) on selectivity parameters (selectivity$re):
- - - + + + @@ -362,7 +362,7 @@ selectivity$re
Now we can run the above models in a loop:
-+for(m in 1:n.mods){ if(sel_model[m] == "logistic"){ # logistic selectivity # overwrite initial parameter values in ASAP data file (ex1_SNEMAYT.dat) @@ -396,11 +396,11 @@
4. Model convergence and comparison
Collect all models into a list:
-+Check which models converged. Note that 3 of the 9 models did NOT converge:
-+vign4_conv <- lapply(mods, function(x) capture.output(check_convergence(x))) for(m in 1:n.mods) cat(paste0("Model ",m,":"), vign4_conv[[m]], "", sep='\n')#> Model 1: @@ -457,7 +457,7 @@#> Max gradient parameter: logit_q #> TMB:sdreport() was performed for this model, but it appears hessian was not invertible
Plot output for models that converged (in a subfolder for each model):
-+# Is Hessian positive definite? ok_sdrep = sapply(mods, function(x) if(x$na_sdrep==FALSE & !is.na(x$na_sdrep)) 1 else 0) pdHess <- as.logical(ok_sdrep) @@ -468,7 +468,7 @@plot_wham_output(mod=mods[[m]], out.type='html', dir.main=file.path(write.dir,paste0("m",m))) }
Compare the models using AIC:
-+df.aic <- as.data.frame(compare_wham_models(mods, sort=FALSE, calc.rho=FALSE)$tab) df.aic[!pdHess,] = NA minAIC <- min(df.aic$AIC, na.rm=T) @@ -502,7 +502,7 @@#> 8 -1477.5 #> 9 NA
Prepare to plot selectivity-at-age for block 1 (fleet).
-+library(tidyverse) library(viridis) @@ -530,7 +530,7 @@df$sel_cor <- factor(df$sel_cor, levels=c("None","IID","AR1","AR1_y","2D AR1")) df$conv = factor(df$conv)
Now plot selectivity-at-age for block 1 (fleet) in all models, with unconverged models pale and converged models solid.
-+-print(ggplot(df, aes(x=Year, y=Age)) + geom_tile(aes(fill=Selectivity, alpha=conv)) + scale_alpha_discrete(range=c(0.4,1), guide=FALSE) + @@ -541,16 +541,18 @@theme_bw() + scale_x_continuous(expand=c(0,0)) + scale_y_continuous(expand=c(0,0)))
+
++
A note on convergence
When fitting age-specific selectivity, oftentimes some of the (mean, \(\mu^{s}_a\)) selectivity parameters need to be fixed for the model to converge. The specifications used here follow this procedure:
-
- Fit the model without fixing any selectivity parameters.
-- If the model fails to converge or the hessian is not invertible (i.e. not positive definite), look for mean selectivity parameters that are very close to 0 or 1 (> 5 or < -5 on the logit scale) and/or have
+NaNestimates of their standard error:- If the model fails to converge or the hessian is not invertible (i.e. not positive definite), look for mean selectivity parameters that are very close to 0 or 1 (> 5 or < -5 on the logit scale) and/or have
NaNestimates of their standard error:+mod$rep$logit_selpars # mean sel pars mod$rep$sel_repars # if time-varying selectivity turned on mod$rep$selAA # selectivity-at-age by block @@ -558,7 +560,7 @@
-
- Re-run the model fixing the worst selectivity-at-age parameter for each block at 0 or 1 as appropriate. In the above age-specific models, we first initialized and fixed ages 4, 4, and 2 for blocks 1-3, respectively. The logit scale selectivity for age 5 and ages 3-4 for blocks 1 and 3 remained around 20, indicating that they too should be fixed at 1. Sometimes just initializing the worst parameter is enough, without fixing it.
+# fix selectivity at 1 for ages 4-5 / 4 / 2-4 in blocks 1-3 input <- prepare_wham_input(asap3, model_name=paste(paste0("Model ",m), sel_model[m], paste(sel_re[[m]], collapse="-"), sep=": "), recruit_model=2, selectivity=list(model=rep("age-specific",3), re=sel_re[[m]], @@ -568,7 +570,7 @@
-
- The goal is to find a set of selectivity parameter initial/fixed values that allow all nested models to converge. Fixing parameters should not affect the NLL much, and any model that is a superset of another should not have a greater NLL (indicates not converged to global minimum). The following commands may be helpful:
+mod.list <- file.path(getwd(),paste0("m",1:n.mods,".rds")) mods <- lapply(mod.list, readRDS) sapply(mods, function(x) check_convergence(x)) diff --git a/docs/articles/ex5_GSI_M.html b/docs/articles/ex5_GSI_M.html index 12579fe5..3d22b2ad 100644 --- a/docs/articles/ex5_GSI_M.html +++ b/docs/articles/ex5_GSI_M.html @@ -156,7 +156,7 @@Ex 5: Time-varying natural mortality linked to the Gulf Stream
- 2D AR1 deviations by age and year (random effects), \(M_{y,a} = M_a + \delta_{y,a} \quad\mathrm{and}\quad \delta_{y,a} \sim \mathcal{N}(0,\Sigma)\)
-We also demonstrate alternate specifications for the link between \(M\) and an environmental covariate, the Gulf Stream Index (GSI), as in O’Leary et al. (2019):
+We also demonstrate alternate specifications for the link between \(M\) and an environmental covariate, the Gulf Stream Index (GSI), as in O'Leary et al. (2019):
- none
- linear (in log-space), \(M_{y,a} = e^{\mathrm{log}\mu_M + \beta_1 E_y}\) @@ -169,37 +169,37 @@
Ex 5: Time-varying natural mortality linked to the Gulf Stream
1. Load data
Open R and load the
-whampackage:+For a clean, runnable
-.Rscript, look atex5_M_GSI.Rin theexample_scriptsfolder of thewhampackage install:+wham.dir <- find.package("wham") file.path(wham.dir, "example_scripts")You can run this entire example script with:
-+-write.dir <- "choose/where/to/save/output" # otherwise will be saved in working directory source(file.path(wham.dir, "example_scripts", "ex5_M_GSI.R"))Let’s create a directory for this analysis:
-+Let's create a directory for this analysis:
+-# choose a location to save output, otherwise will be saved in working directory write.dir <- "choose/where/to/save/output" dir.create(write.dir) setwd(write.dir)We need the same ASAP data file as in example 2, and the environmental covariate (GSI). Let’s copy
-ex2_SNEMAYT.datandGSI.csvto our analysis directory:+We need the same ASAP data file as in example 2, and the environmental covariate (GSI). Let's copy
+ex2_SNEMAYT.datandGSI.csvto our analysis directory:wham.dir <- find.package("wham") file.copy(from=file.path(wham.dir,"extdata","ex2_SNEMAYT.dat"), to=write.dir, overwrite=FALSE) file.copy(from=file.path(wham.dir,"extdata","GSI.csv"), to=write.dir, overwrite=FALSE)Confirm you are in the correct directory and it has the required data files:
-+Read the ASAP3 .dat file into R and convert to input list for wham:
-+asap3 <- read_asap3_dat("ex2_SNEMAYT.dat")Load the environmental covariate (Gulf Stream Index, GSI) data into R:
-+env.dat <- read.csv("GSI.csv", header=T) head(env.dat) #> year GSI @@ -215,7 +215,7 @@
2. Specify models
Now we specify several models with different options for natural mortality:
-+df.mods <- data.frame(M_model = c(rep("---",3),"age-specific","weight-at-age",rep("constant",6),"age-specific","age-specific",rep("constant",3),"---"), M_re = c(rep("none",6),"ar1_y","2dar1","none","none","2dar1","none","2dar1",rep("ar1_a",3),"2dar1"), Ecov_process = rep("ar1",17), @@ -224,7 +224,7 @@df.mods$Model <- paste0("m",1:n.mods) df.mods <- df.mods %>% select(Model, everything()) # moves Model to first col
Look at the model table:
-+df.mods #> Model M_model M_re Ecov_process Ecov_link #> 1 m1 --- none ar1 0 @@ -250,20 +250,13 @@3. Natural mortality options
Next we specify the options for modeling natural mortality by including an optional list argument,
M, to theprepare_wham_input()function (see theprepare_wham_input()help page).Mspecifies estimation options and can overwrite M-at-age values specified in the ASAP data file. By default (i.e.MisNULLor not included), the M-at-age matrix from the ASAP data file is used (M fixed, not estimated).Mis a list with the following entries:-
- -
--
$model: Natural mortality model options.+
-
$model: Natural mortality model options."constant": estimate a single \(M\), shared across all ages and years.- -
"age-specific": estimate \(M_a\) independent for each age, shared across years.- -
-"weight-at-age": estimate \(M\) as a function of weight-at-age, \(M_{y,a} = \mu_M * W_{y,a}^b\), as in Lorenzen (1996) and Miller & Hyun (2018).- -
+-
$re: Time- and age-varying random effects on \(M\).+
-- +
"weight-at-age": estimate \(M\) as a function of weight-at-age, \(M_{y,a} = \mu_M * W_{y,a}^b\), as in Lorenzen (1996) and Miller & Hyun (2018).
$re: Time- and age-varying random effects on \(M\)."none": \(M\) constant in time and across ages (default).- @@ -272,25 +265,22 @@
"ar1_a": \(M\) correlated by age (AR1), constant in time.- -
"ar1_y": \(M\) correlated by year (AR1), constant by age.- -
-"2dar1": \(M\) correlated by year and age (2D AR1), as in Cadigan (2016).
"2dar1": \(M\) correlated by year and age (2D AR1), as in Cadigan (2016).
$initial_means: Initial/mean M-at-age vector, with length = number of ages (if$model = "age-specific") or 1 (if$model = "weight-at-age"or"constant"). IfNULL, initial mean M-at-age values are taken from the first row of the MAA matrix in the ASAP data file.
$est_ages: Vector of ages to estimate age-specific \(M_a\) (others will be fixed at initial values). E.g. in a model with 6 ages,$est_ages = 5:6will estimate \(M_a\) for the 5th and 6th ages, and fix \(M\) for ages 1-4. IfNULL, \(M\) at all ages is fixed atM$initial_means(if notNULL) or row 1 of the MAA matrix from the ASAP file (ifM$initial_means = NULL).
$logb_prior: (Only for weight-at-age model) TRUE or FALSE (default), should a \(\mathcal{N}(0.305, 0.08)\) prior be used onlog_b? Based on Fig. 1 and Table 1 (marine fish) in Lorenzen (1996).For example, to fit model
-m1, fix \(M_a\) at values in ASAP file:+M <- NULL # or simply leave out of call to prepare_wham_inputTo fit model
-m6, estimate one \(M\), constant by year and age:+M <- list(model="constant", est_ages=1)And to fit model
-m8, estimate mean \(M\) with 2D AR1 deviations by year and age:+M <- list(model="constant", est_ages=1, re="2dar1")To fit model
-m17, use the \(M_a\) values specified in the ASAP file, but with 2D AR1 deviations as in Cadigan (2016):+M <-