-
Notifications
You must be signed in to change notification settings - Fork 31
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Doc: Illustrate options for potentially speeding up model import/simulation #1965
Conversation
Codecov Report
Additional details and impacted files@@ Coverage Diff @@
## develop #1965 +/- ##
===========================================
+ Coverage 75.98% 76.02% +0.03%
===========================================
Files 76 76
Lines 12984 12984
===========================================
+ Hits 9866 9871 +5
+ Misses 3118 3113 -5
Flags with carried forward coverage won't be shown. Click here to find out more.
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
👍 nice!
| "\n", | ||
| "#### Parameters as constants\n", | ||
| "\n", | ||
| "By default, AMICI will generate sensitivity equations with respect to all model parameters. If it is clear upfront, that sensitivities with respect to certain parameters will not be required, their IDs can be passed to [amici.sbml_import.SbmlImporter.sbml2amici](https://amici.readthedocs.io/en/latest/generated/amici.sbml_import.SbmlImporter.html#amici.sbml_import.SbmlImporter.sbml2amici) or [amici.pysb_import.pysb2amici](https://amici.readthedocs.io/en/latest/generated/amici.pysb_import.html?highlight=pysb2amici#amici.pysb_import.pysb2amici) via the `constant_parameters` to not generate the respective equations. This will reduce CPU time and RAM requirements during import and simulation.\n", |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
constant_parameters argument
| "#### Parameters as constants\n", | ||
| "\n", | ||
| "By default, AMICI will generate sensitivity equations with respect to all model parameters. If it is clear upfront, that sensitivities with respect to certain parameters will not be required, their IDs can be passed to [amici.sbml_import.SbmlImporter.sbml2amici](https://amici.readthedocs.io/en/latest/generated/amici.sbml_import.SbmlImporter.html#amici.sbml_import.SbmlImporter.sbml2amici) or [amici.pysb_import.pysb2amici](https://amici.readthedocs.io/en/latest/generated/amici.pysb_import.html?highlight=pysb2amici#amici.pysb_import.pysb2amici) via the `constant_parameters` to not generate the respective equations. This will reduce CPU time and RAM requirements during import and simulation.\n", | ||
| "The PEtab import will take care of that automatically.\n", |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
will automatically pass all parameters with petab.ESTIMATE==False as constant_parameters arguments.
| "#### Extracting common subexpressions\n", | ||
| "\n", | ||
| "For some models, the size of the generated model code can be significantly reduced by extracting common subexpressions. This can yield substantial reductions of compile times and RAM-requirements. Very large models might not compile without this option. Extracting common subexpressions can be enabled by setting an environment variable `AMICI_EXTRACT_CSE=1` before model import.\n", | ||
| "The downside is, that the generated model code becomes rather unreadable. The increase in import time when enabling this feature is usually <15%, the effect on code size and compile time is highly model dependent. Mostly models with tightly coupled ODEs, as obtained from complex rate laws or spatial discretizations of ODEs, seem to benefit. For models with mass action kinetics, this option seems to not be helpful and rather increases compile time (e.g., for FröhlichGer2022, the compile time doubles).\n", |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
FröhlichGer2022 is a pysb model, which internally uses expressions/rate laws to avoid reuse of common expressions. Technically also not mass action kinetics due to the thermodynamic model formulation, although the resulting equations are effectively the same.
| "source": [ | ||
| "#### Parallelization\n", | ||
| "\n", | ||
| "For large models or complex model expressions, symbolic computation of the derivatives can be quite time consuming. This can be parallelized by setting the environment variable `AMICI_IMPORT_NPROCS` to the number of parallel processes that should be used. The impact strongly depends on the model. Note that setting this value too may have a negative performance impact (benchmark!).\n", |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
| "For large models or complex model expressions, symbolic computation of the derivatives can be quite time consuming. This can be parallelized by setting the environment variable `AMICI_IMPORT_NPROCS` to the number of parallel processes that should be used. The impact strongly depends on the model. Note that setting this value too may have a negative performance impact (benchmark!).\n", | |
| "For large models or complex model expressions, symbolic computation of the derivatives can be quite time consuming. This can be parallelized by setting the environment variable `AMICI_IMPORT_NPROCS` to the number of parallel processes that should be used. The impact strongly depends on the model. Note that setting this value for smaller model may have a negative performance impact (benchmark!).\n", |
| "\n", | ||
| "If only the objective function gradient is required, adjoint sensitivity analysis are often preferable over forward sensitivity analysis. As a rule of thumb, adjoint sensitivity analysis seem to outperform forward sensitivity analysis for models with more than 20 parameters:\n", | ||
| "\n", | ||
| "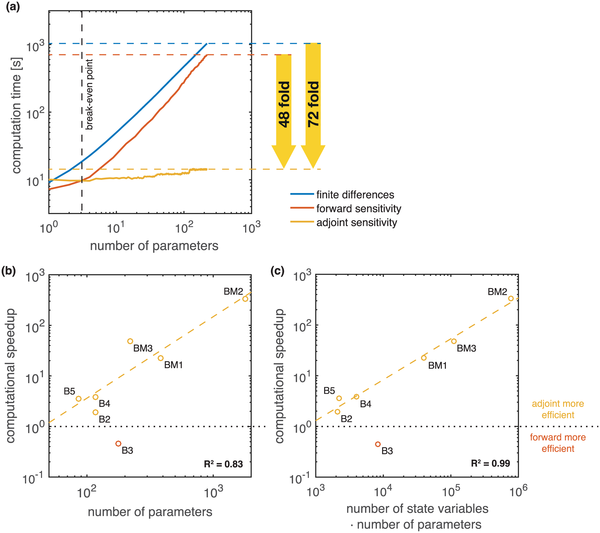\n", |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
looks like we're missing a \n here, not sure why though.
|
Kudos, SonarCloud Quality Gate passed! |








No description provided.