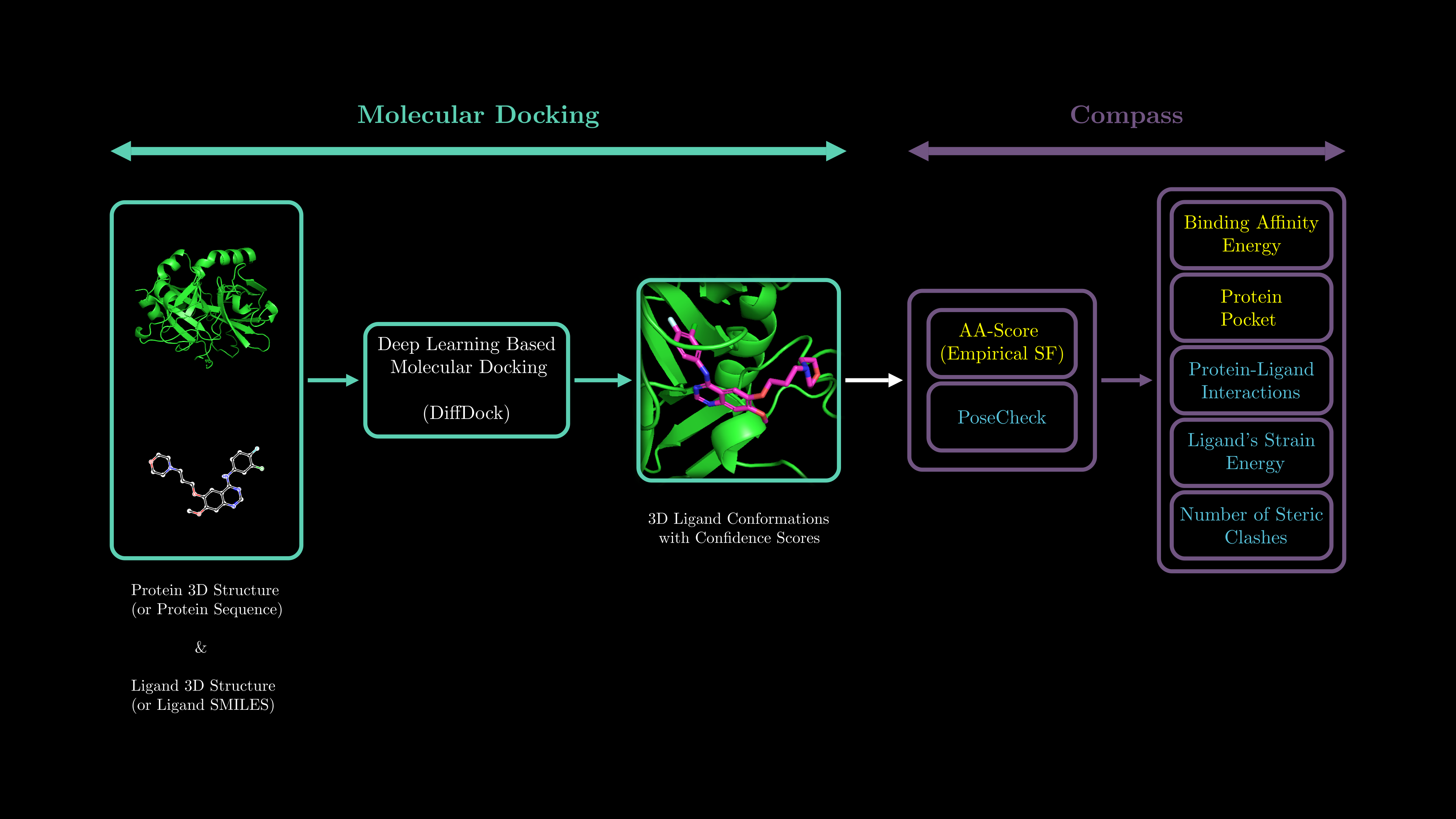
Navigating Future Drugs with CompassDock 🧭
The CompassDock framework is a comprehensive and accurate assessment approach for deep learning-based molecular docking. It evaluates key factors such as the physical and chemical properties, bioactivity favorability of ligands, strain energy, number of protein-ligand steric clashes, binding affinity, and protein-ligand interaction types.
conda create --name CompassDock python=3.11 -c conda-forge
conda install -c ostrokach-forge reduce
conda install -c conda-forge openbabel
conda install -c conda-forge datamol
pip install compassdock
pip install pyg_lib torch_scatter torch_sparse torch_cluster torch_spline_conv -f https://data.pyg.org/whl/torch-2.3.0+cu121.html
pip install "fair-esm @ git+https://github.com/asarigun/esm.git"
pip install "dllogger @ git+https://github.com/NVIDIA/dllogger.git"
pip install "openfold @ git+https://github.com/asarigun/openfold.git"from compassdock import CompassDock
cd = CompassDock()
cd.recursive_compassdocking(
protein_path = 'example/proteins/1a46_protein_processed.pdb',
protein_sequence = None,
ligand_description = 'CCCCC(NC(=O)CCC(=O)O)P(=O)(O)OC1=CC=CC=C1',
complex_name = 'complex_1',
molecule_name = 'molecule_1',
out_dir = 'results',
compass_all = False)
results = cd.results()
print(results)from compassdock import CompassDock
# Initialize CompassDock
cd = CompassDock()
# Perform docking using the provided protein and ligand information
cd.recursive_compassdocking(
protein_path = None,
protein_sequence = 'GIQSYCTPPYSVLQDPPQPVV',
ligand_description = 'CCCCC(NC(=O)CCC(=O)O)P(=O)(O)OC1=CC=CC=C1',
complex_name = 'complex_1',
molecule_name = 'molecule_1',
out_dir = 'results',
compass_all = False,
max_redocking_step = 1)
# Retrieve and print the docking results
results = cd.results()
print(results)More examples can be found in 
For instructions on how to use Fine-Tuning Mode, please refer to the previous branch
Please cite the following paper if you use this code/repository in your research:
@article{sarigun2024compass,
title={CompassDock: Comprehensive Accurate Assessment Approach for Deep Learning-Based Molecular Docking in Inference and Fine-Tuning},
author={Sarigun, Ahmet and Franke, Vedran and Uyar, Bora and Akalin, Altuna},
journal={arXiv preprint arXiv:2406.06841},
year={2024}
}
We extend our deepest gratitude to the following teams for open-sourcing their valuable Repos:
- DiffDock Team (version 2023 & 2024),
- AA-score Team,
- PoseCheck Team