Akarsh Kumar
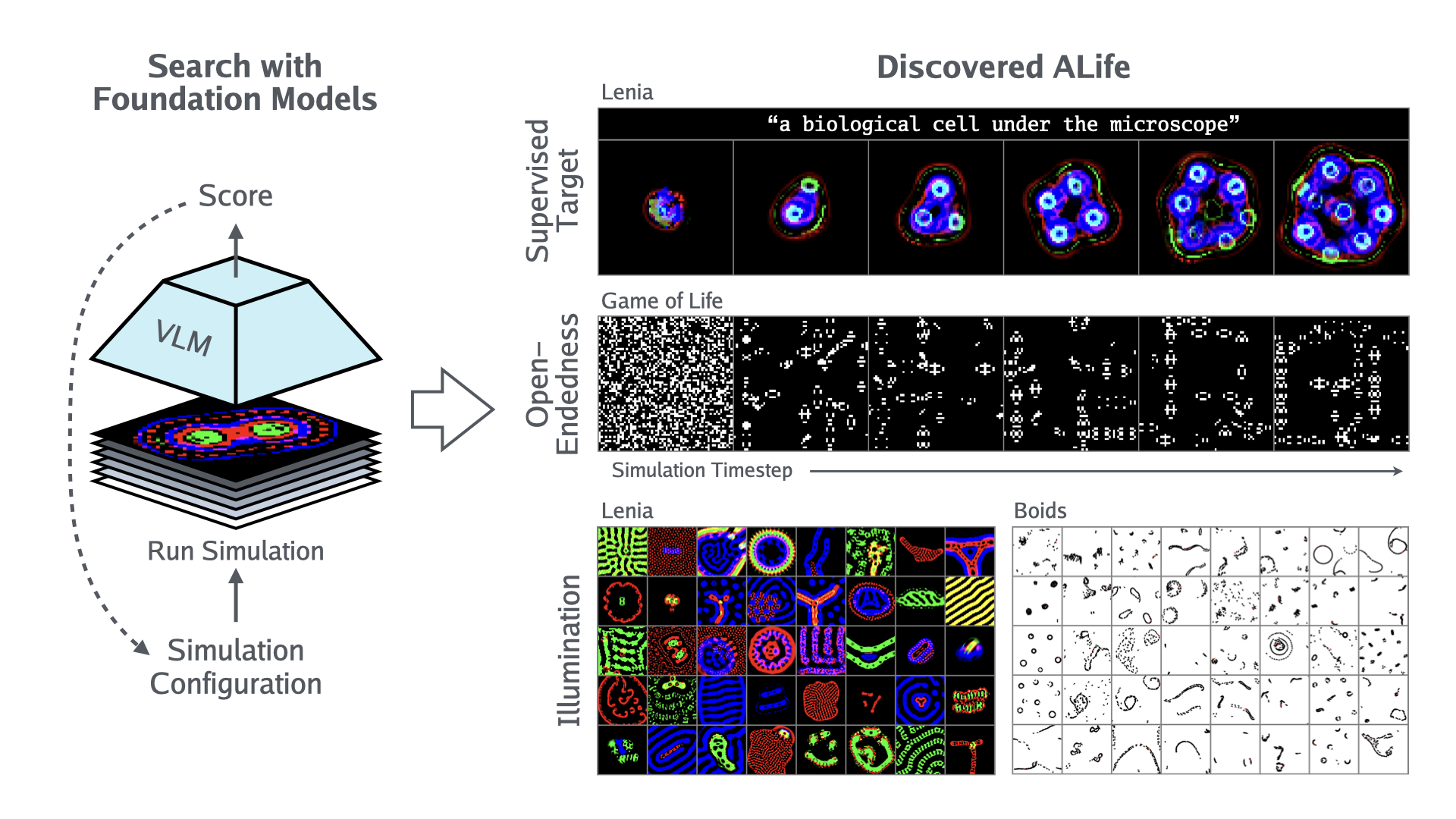
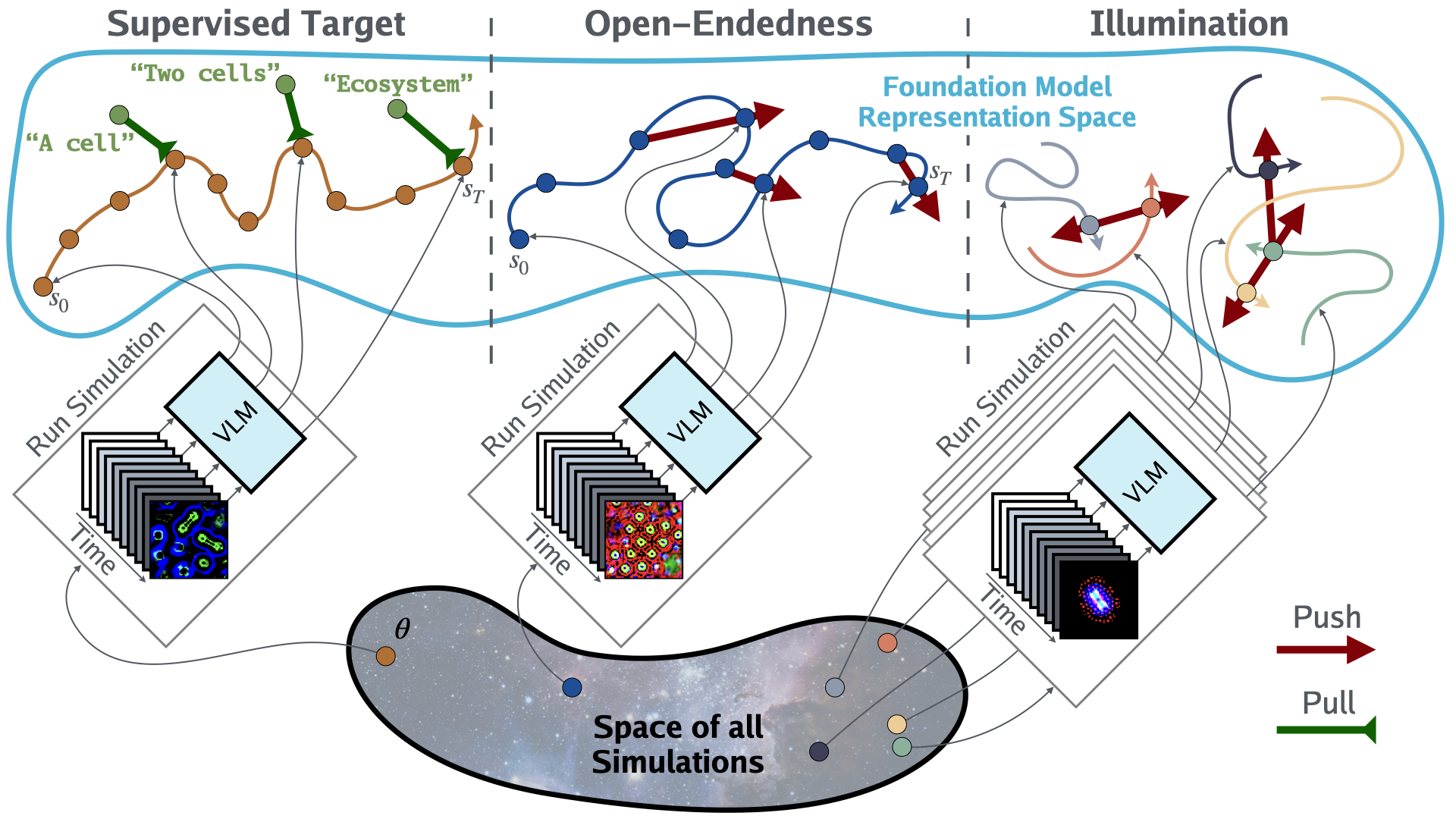
With the recent Nobel Prize awarded for radical advances in protein discovery, foundation models (FMs) for exploring large combinatorial spaces promise to revolutionize many scientific fields. Artificial Life (ALife) has not yet integrated FMs, thus presenting a major opportunity for the field to alleviate the historical burden of relying chiefly on manual design and trial-and-error to discover the configurations of lifelike simulations. This paper presents, for the first time, a successful realization of this opportunity using vision-language FMs. The proposed approach, called Automated Search for Artificial Life (ASAL), (1) finds simulations that produce target phenomena, (2) discovers simulations that generate temporally open-ended novelty, and (3) illuminates an entire space of interestingly diverse simulations. Because of the generality of FMs, ASAL works effectively across a diverse range of ALife substrates including Boids, Particle Life, Game of Life, Lenia, and Neural Cellular Automata. A major result highlighting the potential of this technique is the discovery of previously unseen Lenia and Boids lifeforms, as well as cellular automata that are open-ended like Conway’s Game of Life. Additionally, the use of FMs allows for the quantification of previously qualitative phenomena in a human-aligned way. This new paradigm promises to accelerate ALife research beyond what is possible through human ingenuity alone.
This repo contains a minimalistic implementation of ASAL to get you started ASAP. Everything is implemented in the Jax framework, making everything end-to-end jittable and very fast.
The important code is here:
- foundation_models/init.py has the code to create a foundation model.
- substrates/init.py has the code to create a substrate.
- rollout.py has the code to rollout a simulation efficiently.
- asal_metrics.py has the code to compute the metrics from ASAL.
Here is some minimal code to sample some random simulation parameters and run the simulation and evaluate how open-ended it is:
import jax
from functools import partial
import substrates
import foundation_models
from rollout import rollout_simulation
import asal_metrics
fm = foundation_models.create_foundation_model('clip')
substrate = substrates.create_substrate('lenia')
rollout_fn = partial(rollout_simulation, s0=None, substrate=substrate, fm=fm, rollout_steps=substrate.rollout_steps, time_sampling=8, img_size=224, return_state=False) # create the rollout function
rollout_fn = jax.jit(rollout_fn) # jit for speed
# now you can use rollout_fn as you need...
rng = jax.random.PRNGKey(0)
params = substrate.default_params(rng) # sample random parameters
rollout_data = rollout_fn(rng, params)
rgb = rollout_data['rgb'] # shape: (8, 224, 224, 3)
z = rollout_data['z'] # shape: (8, 512)
oe_score = asal_metrics.calc_open_endedness_score(z) # shape: ()We have already implemented the following ALife substrates:
- 'lenia': Lenia
- 'boids': Boids
- 'plife': Particle Life
- 'plife_plus': Particle Life++
- (Particle Life with changing color dynamics)
- 'plenia': Particle Lenia
- 'dnca': Discrete Neural Cellular Automata
- 'nca_d1': Continuous Neural Cellular Automata
- 'gol': Game of Life/Life-Like Cellular Automata
You can find the code for these substrates at substrates/
The main files to run the entire ASAL pipeline are the following:
- main_opt.py
- Run this for supervised target and open-endedness
- Search algorithm: Sep-CMA-ES (from evosax)
- main_illuminate.py
- Run this for illumination
- Search algorithm: custom genetic algorithm
- main_sweep_gol.py
- Run this for open-endedness in Game of Life substrate (b/c discrete search space)
- Search algorithm: brute force search
asal.ipynb goes through everything you need to know.
Check out the Google Colab here!
To run this project locally, you can start by cloning this repo.
git clone https://github.com/SakanaAI/asal.gitThen, set up the python environment with conda:
conda create --name asal python=3.10.13
conda activate asalThen, install the necessary python libraries:
python -m pip install -r requirements.txtHowever, if you want GPU acceleration (trust me, you do), please manually install jax according to your system's CUDA version.
Check out asal.ipynb to learn how to run the files and visualize the results.
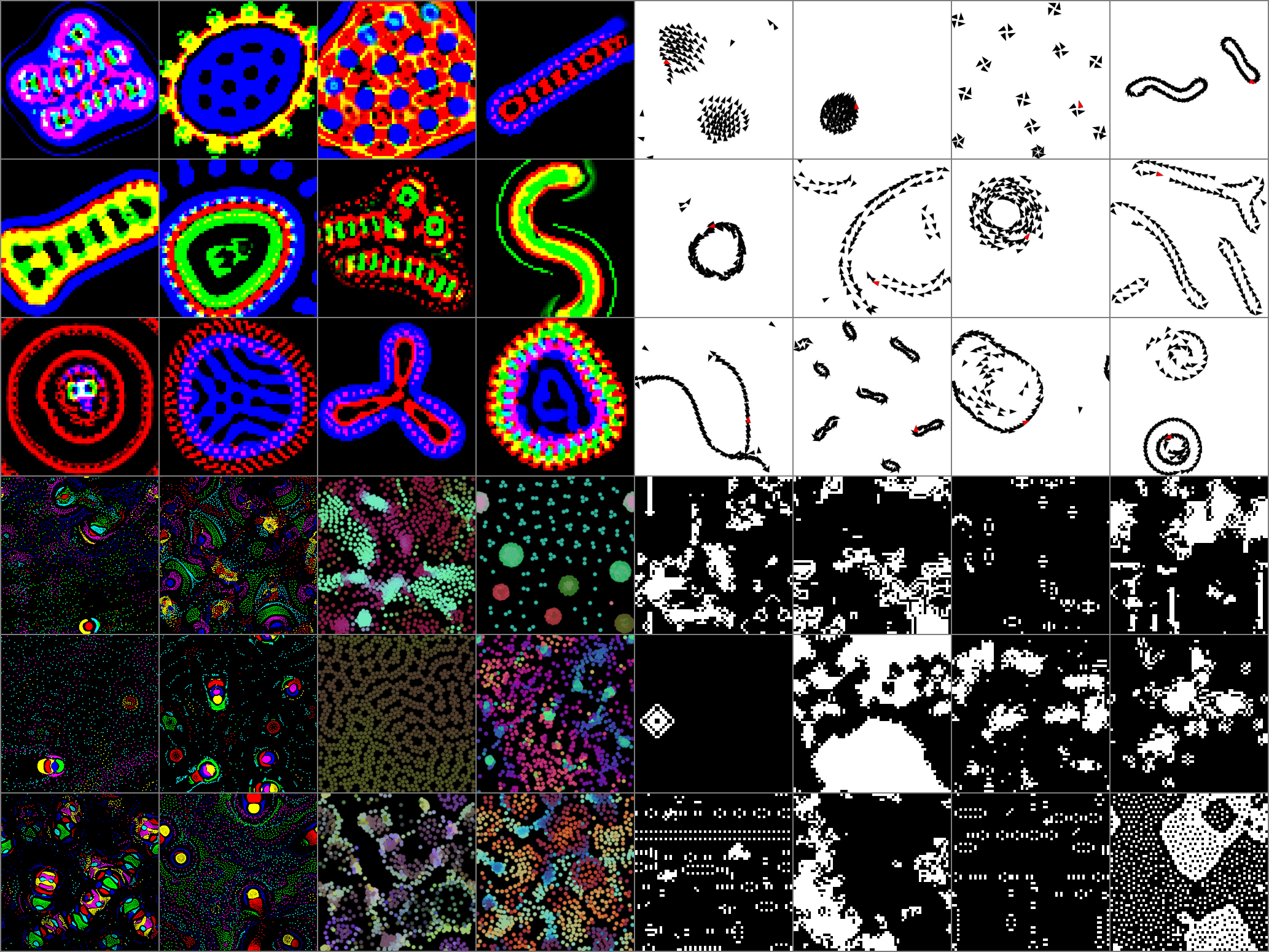
You can view our dataset of simulations at
- https://pub.sakana.ai/asal/data/illumination_poster_lenia.png
- https://pub.sakana.ai/asal/data/illumination_poster_boids.png and find the simulation parameters at
- https://pub.sakana.ai/asal/data/illumination_lenia.npz
- https://pub.sakana.ai/asal/data/illumination_boids.npz
- https://pub.sakana.ai/asal/data/illumination_plife.npz
- https://pub.sakana.ai/asal/data/sweep_gol.npz
Directions on how to load these simulations are shown in asal.ipynb.
Everything you need is already in this repo.
If, for some reason, you want to see more code and see what went into the experimentation that led to the creation of ASAL, then check out this repo.
To cite our work, you can use the following:
@article{kumar2024asal,
title = {Automating the Search for Artificial Life with Foundation Models},
author = {Akarsh Kumar and Chris Lu and Louis Kirsch and Yujin Tang and Kenneth O. Stanley and Phillip Isola and David Ha},
year = {2024},
url = {https://asal.sakana.ai/}
}