Install (or upgrade) memory_graph using pip:
pip install --upgrade memory_graph
Additionally Graphviz needs to be installed.
In Python, assigning the list from variable a to variable b causes both variables to reference the same list object and therefore share the data. Consequently, any change applied through one variable will impact the other. This behavior can lead to elusive bugs if a programmer incorrectly assumes that list a and b are independent.
import memory_graph as mg
# create the lists 'a' and 'b'
a = [4, 3, 2]
b = a
a.append(1) # changing 'a' changes 'b'
# print the 'a' and 'b' list
print('a:', a)
print('b:', b)
# check if 'a' and 'b' share data
print('ids:', id(a), id(b))
print('identical?:', a is b)
# show all local variables in a graph
mg.show( locals() ) |
a graph showing |
The fact that a and b share data can not be verified by printing the lists. It can be verified by comparing the identity of both variables using the id() function or by using the is comparison operator as shown in the program output below, but this quickly becomes impractical for larger programs.
a: 4, 3, 2, 1
b: 4, 3, 2, 1
ids: 126432214913216 126432214913216
identical?: True
A better way to understand what data is shared is to draw a graph of the data using the memory_graph package.
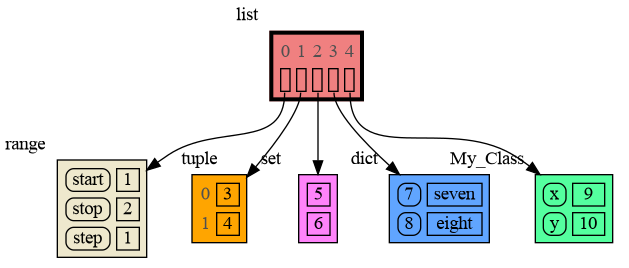
The memory_graph package can graph many different data types, not limited to:
import memory_graph as mg
class MyClass:
def __init__(self, x, y):
self.x = x
self.y = y
data = [ range(1, 2), (3, 4), {5, 6}, {7:'seven', 8:'eight'}, MyClass(9, 10) ]
mg.show(data)Instead of showing the graph you can also render it to an output file of your choosing (see Graphviz Output Formats) using for example:
mg.render(data, "my_graph.pdf")
mg.render(data, "my_graph.svg")
mg.render(data, "my_graph.png")
mg.render(data, "my_graph.gv") # Graphviz DOT file
mg.render(data) # renders to 'mg.render_filename' with default value: 'memory_graph.pdf'Bas Terwijn
Inspired by Python Tutor.
The Python Data Model makes a distiction between immutable and mutable types:
- immutable: bool, int, float, complex, str, tuple, bytes, frozenset
- mutable: list, set, dict, classes, ... (most other types)
In the code below variable a and b both reference the same tuple value (4, 3, 2). A tuple is an immutable type and therefore when we change variable a its value cannot be mutated in place, and thus a copy is made and a and b reference a different value afterwards.
import memory_graph as mg
a = (4, 3, 2)
b = a
mg.render(locals(), 'immutable1.png')
a += (1,)
mg.render(locals(), 'immutable2.png')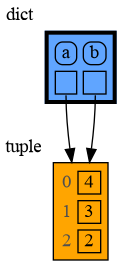 |
 |
|---|---|
| immutable1.png | immutable2.png |
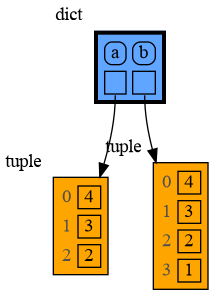
With mutable types the result is different. In the code below variable a and b both reference the same list value [4, 3, 2]. A list is a mutable type and therefore when we change variable a its value can be mutated in place and thus a and b both reference the same new value afterwards. Thus changing a also changes b and vice versa. Sometimes we want this but other times we don't and then we will have to make a copy ourselfs so that a and b are independent.
import memory_graph as mg
a = [4, 3, 2]
b = a
mg.render(locals(), 'mutable1.png')
a += [1] # equivalent to: a.append(1)
mg.render(locals(), 'mutable2.png')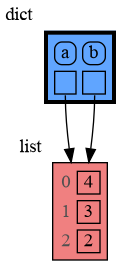 |
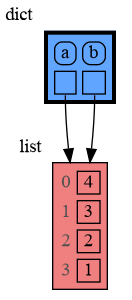 |
|---|---|
| mutable1.png | mutable2.png |
One practical reason why Python makes the distinction between mutable and immutable types is that a value of a mutable type can be large, making it inefficient to copy each time we change it. Immutable values generally don't need to change as much, or are small making copying less of a concern.
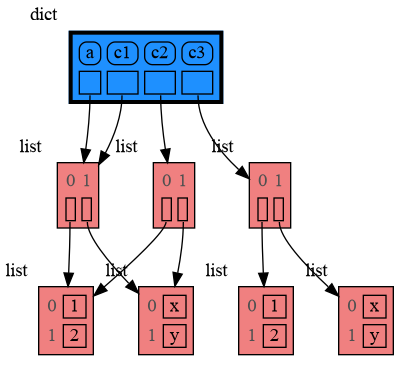
Python offers three different "copy" options that we will demonstrate using a nested list:
import memory_graph as mg
import copy
a = [ [1, 2], ['x', 'y'] ] # a nested list (a list containing lists)
# three different ways to make a "copy" of 'a':
c1 = a
c2 = copy.copy(a) # equivalent to: a.copy() a[:] list(a)
c3 = copy.deepcopy(a)
mg.show(locals())c1is an assignment, nothing is copied, all the values are sharedc2is a shallow copy, only the value referenced by the first reference is copied, all the underlying values are sharedc3is a deep copy, all the values are copied, nothing is shared
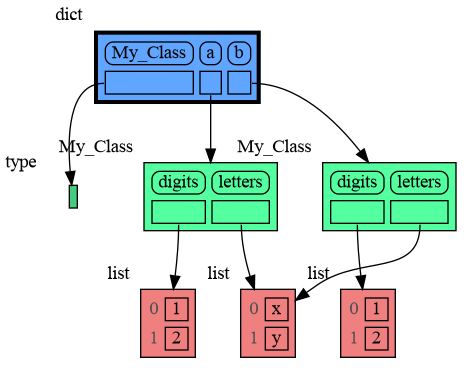
We can write our own custom copy function or method in case the three "copy" options don't do what we want. For example, in the code below the copy() method of My_Class copies the digits but shares the letters between two objects.
import memory_graph as mg
import copy
class My_Class:
def __init__(self):
self.digits = [1, 2]
self.letters = ['x', 'y']
def copy(self): # custom copy method copies the digits but shares the letters
c = copy.copy(self)
c.digits = copy.copy(self.digits)
return c
a = My_Class()
b = a.copy()
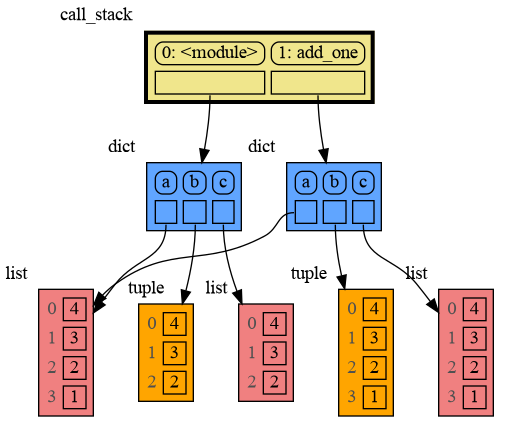
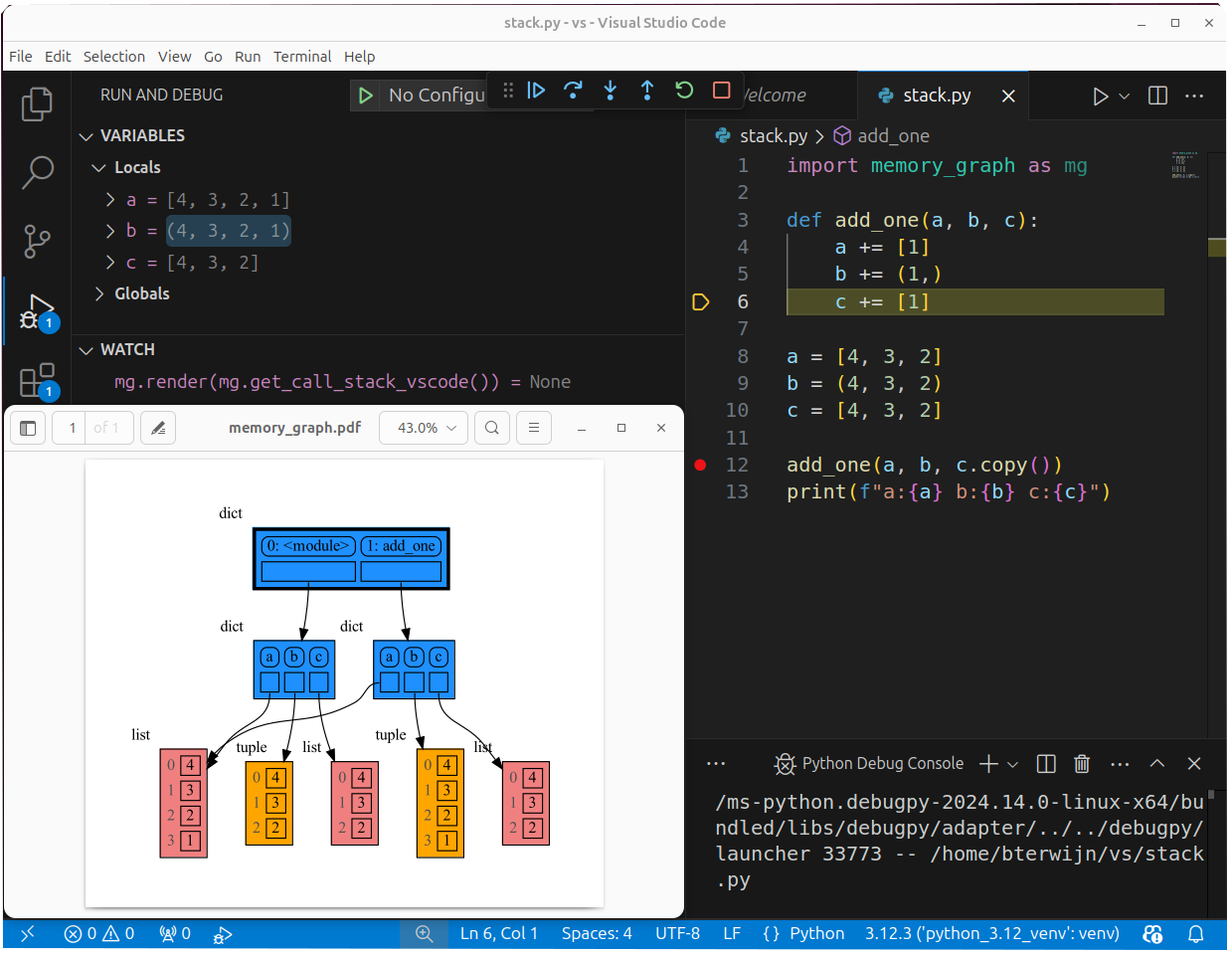
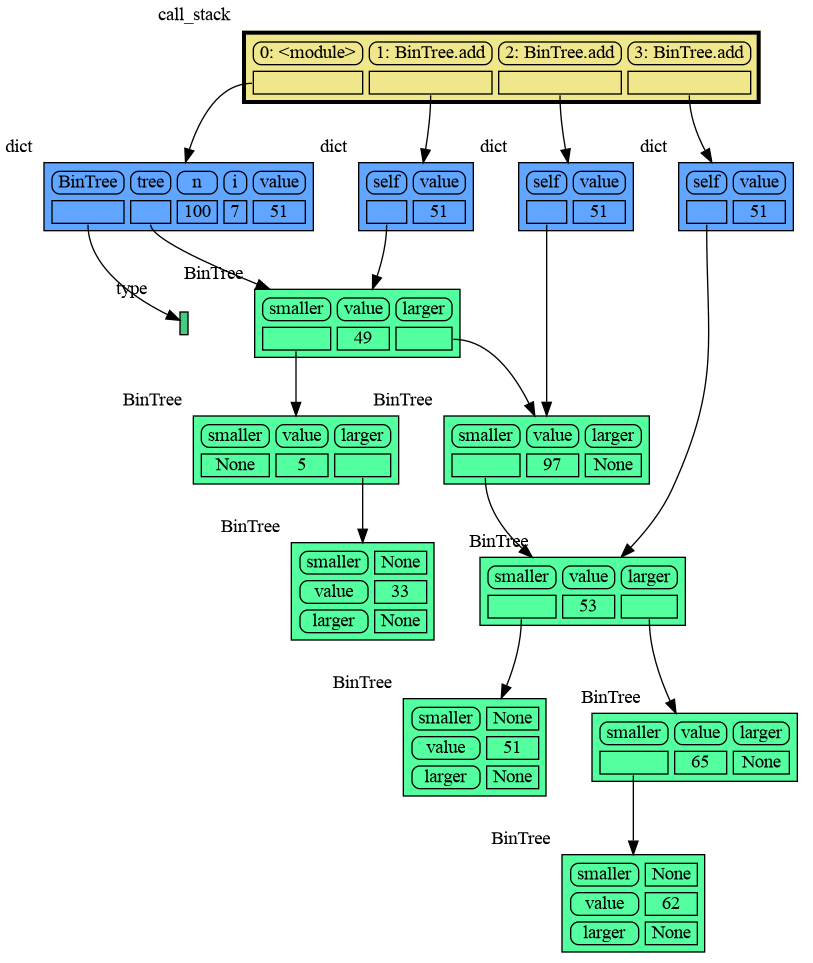
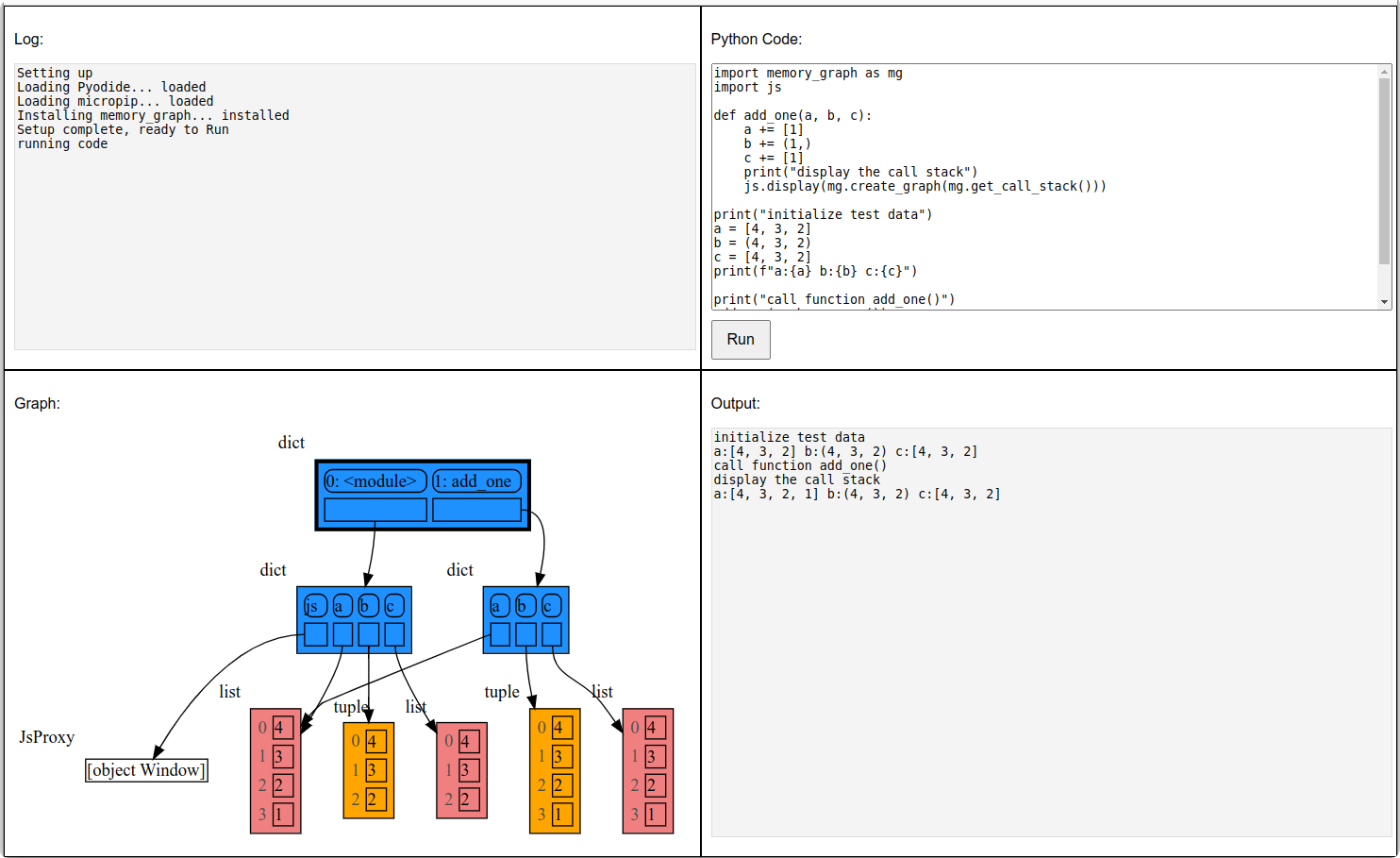
mg.show(locals())The mg.get_call_stack() function retrieves the entire call stack, including the local variables for each function on the stack. This enables us to visualize the local variables across all active functions simultaneously. Then by examining the graph, we can determine whether any local variables from different functions on the call stack share data. For instance, consider the function add_one() which adds the value 1 to each of its parameters a, b, and c.
import memory_graph as mg
def add_one(a, b, c):
a += [1]
b += (1,)
c += [1]
mg.show(mg.get_call_stack())
a = [4, 3, 2]
b = (4, 3, 2)
c = [4, 3, 2]
add_one(a, b, c.copy())
print(f"a:{a} b:{b} c:{c}")In the printed output only a is changed as a result of the function call:
a:[4, 3, 2, 1] b:(4, 3, 2) c:[4, 3, 2]
This is because b is of immutable type 'tuple' so its value gets copied automatically when it is changed. And because the function is called with a copy of c, its original value is not changed by the function. The value of variable a is the only value of mutable type that is shared between the root stack frame '0: <module>' and the '1: add_one' stack frame of the function so only that variable is affected as a result of the function call. The other changes remain confined to the local variables of the add_one() function.
It is often helpful to temporarily block program execution to inspect the graph. For this, you can use the mg.block() function:
mg.block(fun, arg1, arg2, ...) This function:
- first executes
fun(arg1, arg2, ...) - then prints the current source location in the program
- then blocks execution until the <Enter> key is pressed
- finally returns the value of the
fun()call
to change its behavior:
- Set
mg.block_prints_location = Falseto skip printing the source location. - Set
mg.press_enter_message = Noneto skip printing "Press <Enter> to continue...".
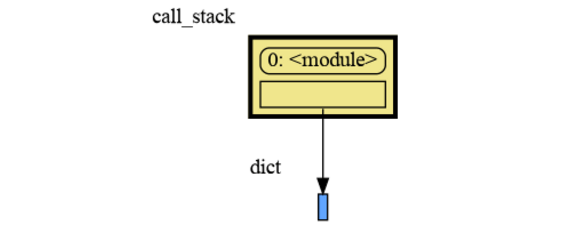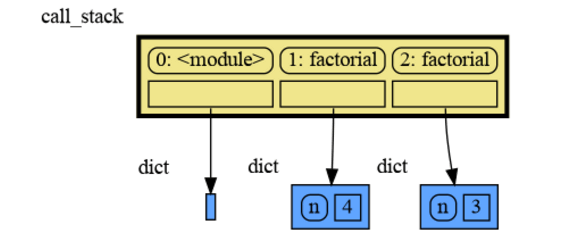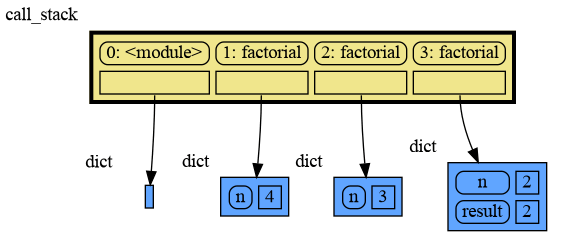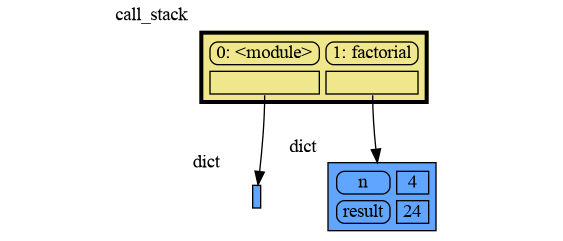
The call stack is also helpful to visualize how recursion works. Here we use mg.block() to show each step of how recursively factorial(3) is computed:
import memory_graph as mg
def factorial(n):
if n==0:
return 1
mg.block(mg.show, mg.get_call_stack())
result = n * factorial(n-1)
mg.block(mg.show, mg.get_call_stack())
return result
print(factorial(3))and the result is: 1 x 2 x 3 = 6
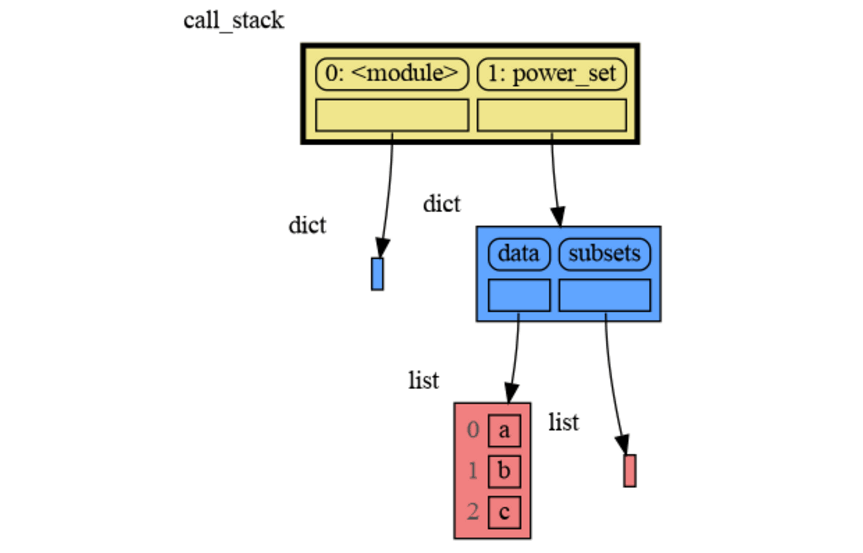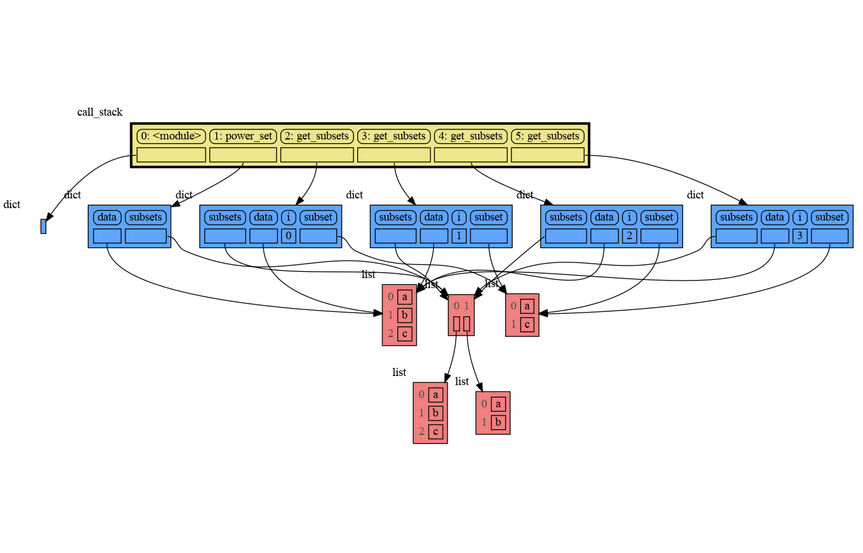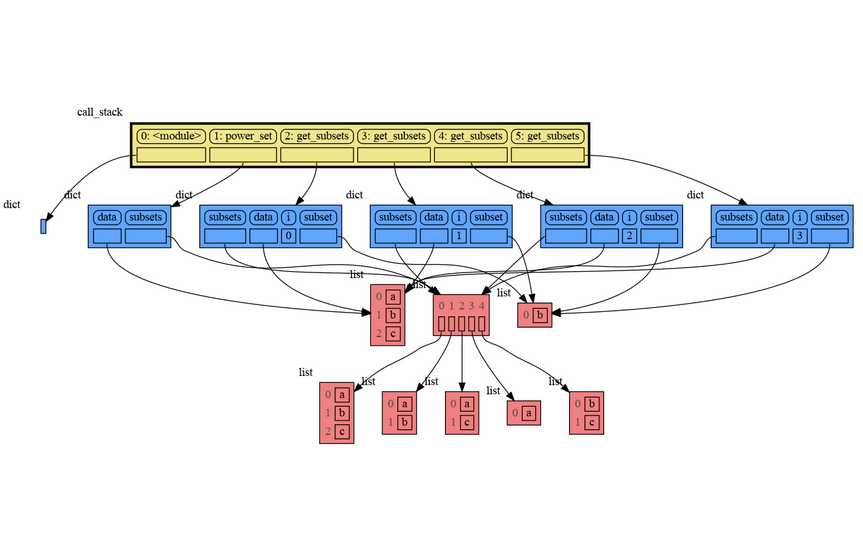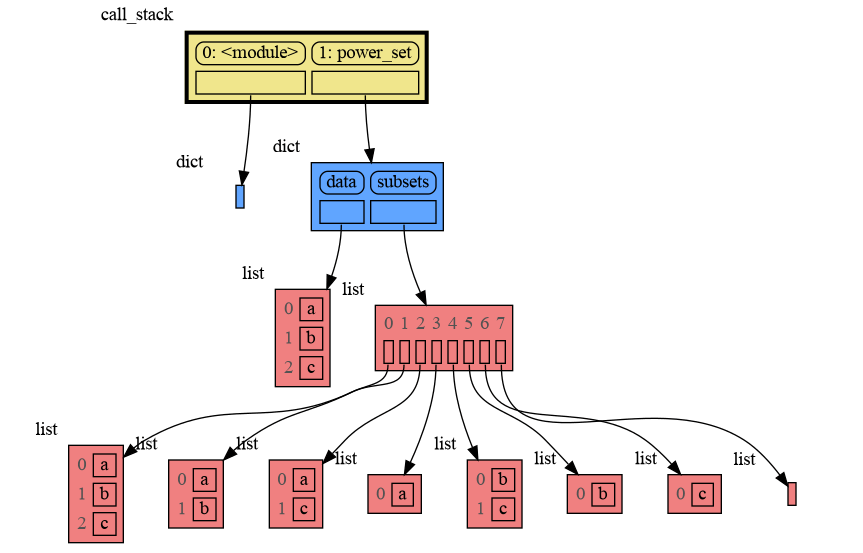
A more interesting recursive example that shows sharing of data is power_set(). A power set is the set of all subsets of a collection of values.
import memory_graph as mg
def get_subsets(subsets, data, i, subset):
mg.block(mg.show, mg.get_call_stack())
if i == len(data):
subsets.append(subset.copy())
return
subset.append(data[i])
get_subsets(subsets, data, i+1, subset) # do include data[i]
subset.pop()
get_subsets(subsets, data, i+1, subset) # don't include data[i]
mg.block(mg.show, mg.get_call_stack())
def power_set(data):
subsets = []
get_subsets(subsets, data, 0, [])
return subsets
print( power_set(['a', 'b', 'c']) )[['a', 'b', 'c'], ['a', 'b'], ['a', 'c'], ['a'], ['b', 'c'], ['b'], ['c'], []]
For the best debugging experience with memory_graph set for example expression:
mg.render(locals(), "my_graph.pdf")
as a watch in a debugger tool such as the integrated debugger in Visual Studio Code. Then open the "my_graph.pdf" output file to continuously see all the local variables while debugging. This avoids having to add any memory_graph show() or render() calls to your code.
The mg.get_call_stack() doesn't work well in watch context in most debuggers because debuggers introduce additional stack frames that cause problems. Use these alternative functions for various debuggers to filter out these problematic stack frames:
| debugger | function to get the call stack |
|---|---|
| pdb, pudb | mg.get_call_stack_pdb() |
| Visual Studio Code | mg.get_call_stack_vscode() |
| Pycharm | mg.get_call_stack_pycharm() |
For other debuggers, invoke this function within the watch context. Then, in the "call_stack.txt" file, identify the slice of functions you wish to include in the call stack.
mg.save_call_stack("call_stack.txt")
Choose 'after' and 'up_to' what function you want to slice and then call this function to get the desired call stack:
mg.get_call_stack_after_up_to(after_function, up_to_function="<module>")
To simplify debugging without a debugger tool, we offer these alias functions that you can insert into your code at the point where you want to visualize a graph:
| alias | purpose | function call |
|---|---|---|
mg.sl() |
show local variables | mg.show(locals()) |
mg.ss() |
show the call stack | mg.show(mg.get_call_stack()) |
mg.bsl() |
block after showing local variables | mg.block(mg.show, locals()) |
mg.bss() |
block after showing the call stack | mg.block(mg.show, mg.get_call_stack()) |
mg.rl() |
render local variables | mg.render(locals()) |
mg.rs() |
render the call stack | mg.render(mg.get_call_stack()) |
mg.brl() |
block after rendering local variables | mg.block(mg.render, locals()) |
mg.brs() |
block after rendering the call stack | mg.block(mg.render, mg.get_call_stack()) |
mg.l() |
same as mg.bsl() |
|
mg.s() |
same as mg.bss() |
For example, executing this program:
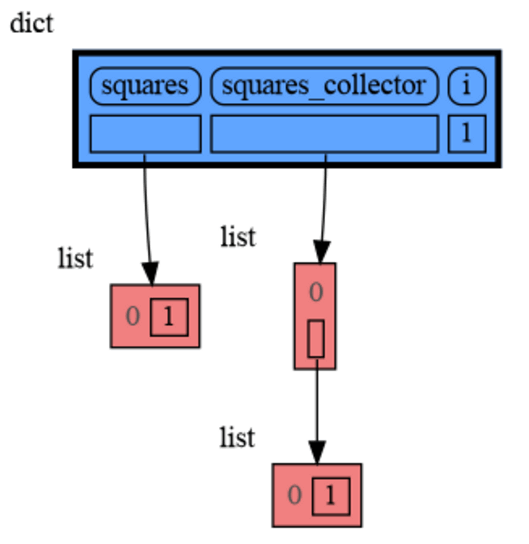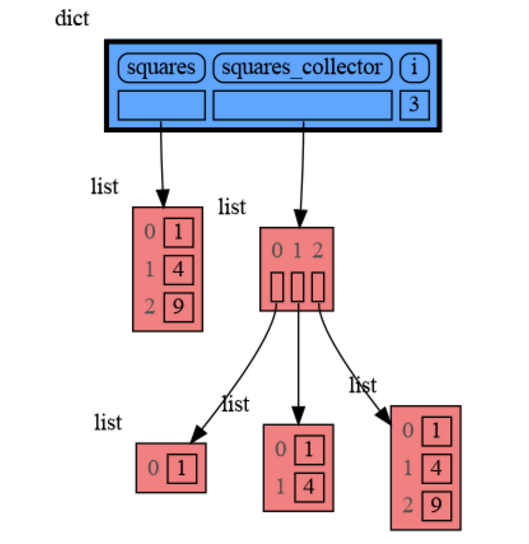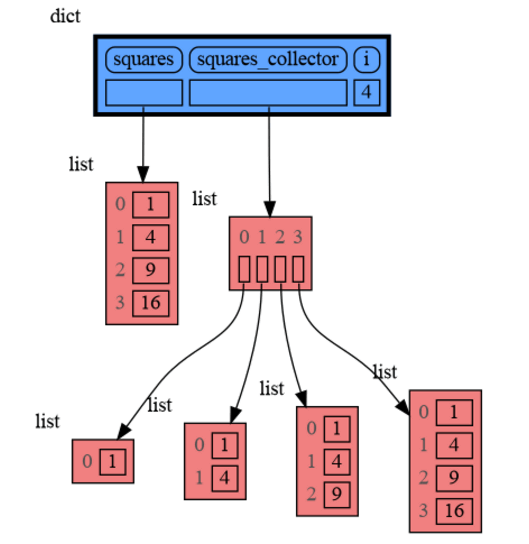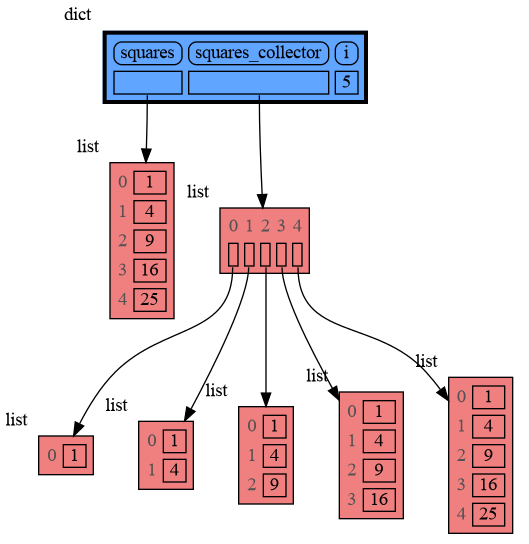
from memory_graph as mg
squares = []
squares_collector = []
for i in range(1, 6):
squares.append(i**2)
squares_collector.append(squares.copy())
mg.l() # block after showing local variablesand pressing <Enter> a number of times, results in:
Module memory_graph can be very useful in a course about datastructures, some examples:
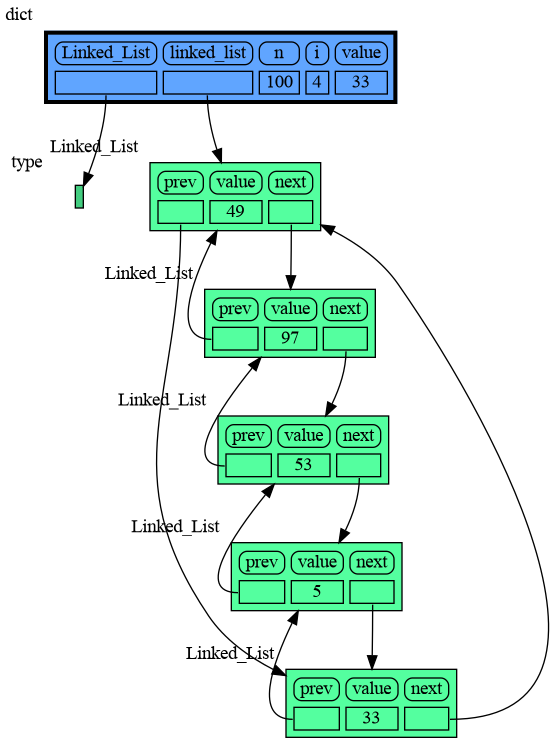
import memory_graph as mg
import random
random.seed(0) # use same random numbers each run
class Node:
def __init__(self, value):
self.prev = None
self.value = value
self.next = None
class LinkedList:
def __init__(self):
self.head = None
self.tail = None
def add_front(self, value):
new_node = Node(value)
if self.head is None:
self.head = new_node
self.tail = new_node
else:
new_node.next = self.head
self.head.prev = new_node
self.head = new_node
mg.block(mg.show, locals()) # <--- draw graph
linked_list = LinkedList()
n = 100
for i in range(n):
new_value = random.randrange(n)
linked_list.add_front(new_value)import memory_graph as mg
import random
random.seed(0) # use same random numbers each run
class Node:
def __init__(self, value):
self.smaller = None
self.value = value
self.larger = None
class BinTree:
def __init__(self):
self.root = None
def add_recursive(self, new_value, node):
if new_value < node.value:
if node.smaller is None:
node.smaller = Node(new_value)
else:
self.add_recursive(new_value, node.smaller)
else:
if node.larger is None:
node.larger = Node(new_value)
else:
self.add_recursive(new_value, node.larger)
mg.block(mg.show, locals()) # <--- draw graph
def add(self, value):
if self.root is None:
self.root = Node(value)
else:
self.add_recursive(value, self.root)
tree = BinTree()
n = 100
for i in range(n):
new_value = random.randrange(100)
tree.add(new_value)import memory_graph as mg
import random
random.seed(0) # use same random numbers each run
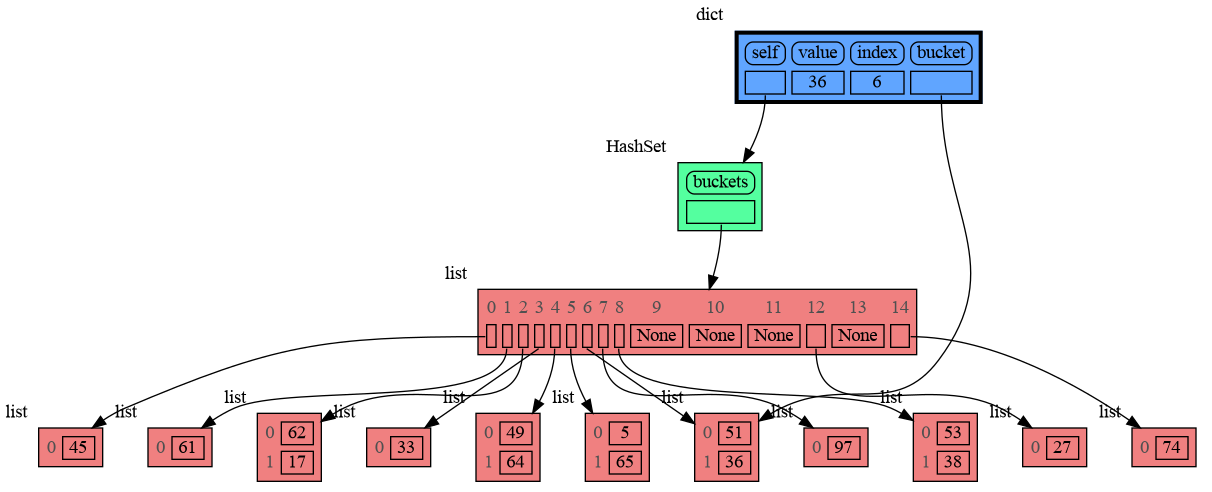
class HashSet:
def __init__(self, capacity=15):
self.buckets = [None] * capacity
def add(self, value):
index = hash(value) % len(self.buckets)
if self.buckets[index] is None:
self.buckets[index] = []
bucket = self.buckets[index]
bucket.append(value)
mg.block(mg.show, locals()) # <--- draw graph
def contains(self, value):
index = hash(value) % len(self.buckets)
if self.buckets[index] is None:
return False
return value in self.buckets[index]
def remove(self, value):
index = hash(value) % len(self.buckets)
if self.buckets[index] is not None:
self.buckets[index].remove(value)
hash_set = HashSet()
n = 100
for i in range(n):
new_value = random.randrange(n)
hash_set.add(new_value)Different aspects of memory_graph can be configured. The default configuration is reset by importing 'memory_graph.config_default'.
-
mg.config.max_graph_depth : int
- The maxium depth of the graph with default value 12. A
✂(scissor) symbol indicates where the graph is cut short. Dashed references indicate that there are more references to a node than are shown.
- The maxium depth of the graph with default value 12. A
-
mg.config.max_string_length : int
- The maximum length of strings shown in the graph. Longer strings will be truncated.
-
mg.config.not_node_types : set
- Holds all types for which no seperate node is drawn but that instead are shown as elements in their parent Node.
-
mg.config.no_child_references_types : set
- The set of key_value types that don't draw references to their direct childeren but have their children shown as elements of their node.
-
mg.config.type_to_node : dict
- Determines how a data types is converted to a Node (sub)class for visualization in the graph.
-
mg.config.type_to_color : dict
- Maps each type to the graphviz color it gets in the graph.
-
mg.config.type_to_vertical_orientation : dict
- Maps each type to its orientation. Use 'True' for vertical and 'False' for horizontal. If not specified Node_Linear and Node_Key_Value are vertical unless they have references to children.
-
mg.config.type_to_slicer : dict
- Maps each type to a Slicer. A slicer determines how many elements of a data type are shown in the graph to prevent the graph from getting too big. 'Slicer()' does no slicing, 'Slicer(1,2,3)' shows just 1 element at the beginning, 2 in the middle, and 3 at the end.
Memory_graph simplifies the visualization (and the viewer's mental model) by not showing separate nodes for immutable types like bool, int, float, complex, and str by default. This simplification can sometimes be slightly misleading. As in the example below, after a shallow copy, lists a and b technically share their int values, but the graph makes it appear as though a and b each have their own copies. However, since int is immutable, this simplification will never lead to unexpected changes or bugs—changing a won’t effect b.
The simplification strikes a balance: it is slightly misleading but keeps the graph clean and easy to understand and focuses on the mutable types where unexpected changes can occur. This is why it is the default behavior. If you do want to show separate nodes for int values, such as for educational purposes, you can simply remove int from the mg.config.not_node_types set:
import memory_graph as mg
a = [100, 200, 300]
b = a.copy()
mg.render(locals(), 'not_node_types1.png')
mg.config.not_node_types.remove(int) # now show separate nodes for int values
mg.render(locals(), 'not_node_types2.png') |
 |
|---|---|
| not_node_types1.png, simplified | not_node_types2.png, technically correct |
Additionally, the simplification hides the reuse of small int values in the current CPython implementation, an optimization that might otherwise confuse beginner Python programmers. For instance, after executing a[1]+=1; b[1]+=1, the 201 value is, maybe surprisingly, still shared between a and b, whereas executing a[2]+=1; b[2]+=1 does not result in sharing the 301 value.
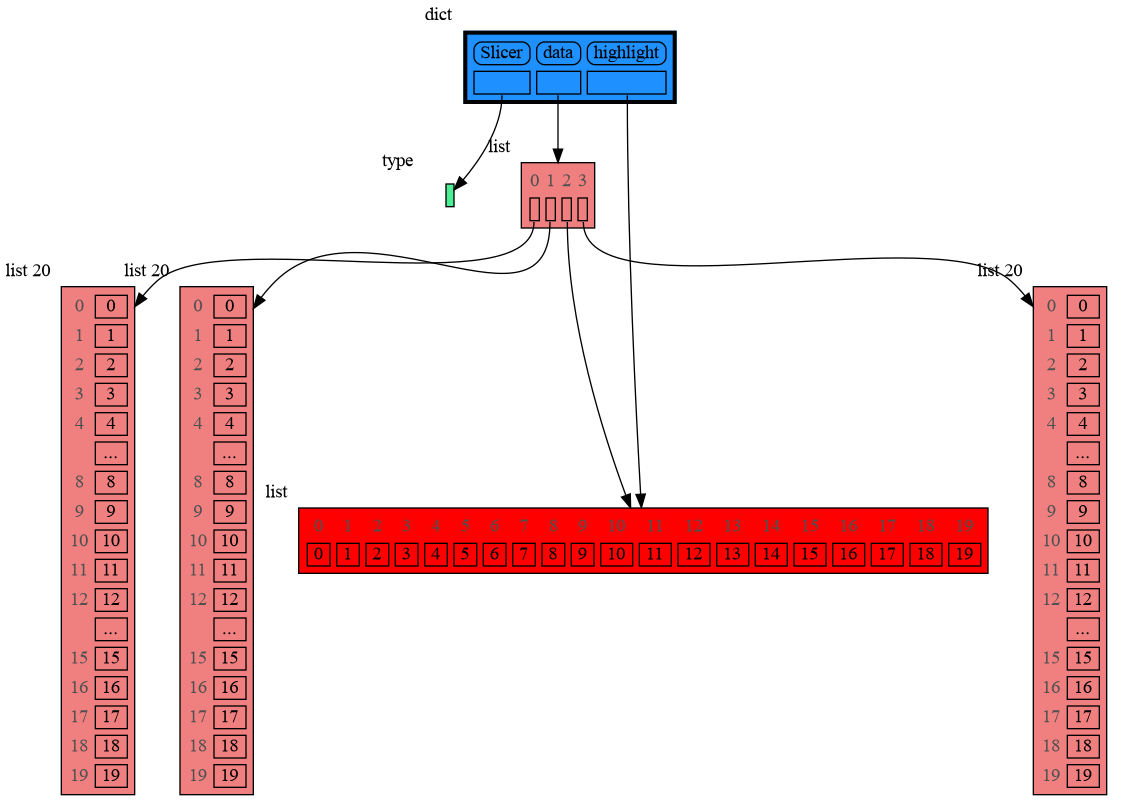
In addition to the global configuration, a temporary configuration can be set for a single show() or render() call to change the colors, orientation, and slicer. This example highlights a particular list element in red, gives it a horizontal orientation, and overwrites the default slicer for lists:
import memory_graph as mg
from memory_graph.slicer import Slicer
data = [ list(range(20)) for i in range(1,5)]
highlight = data[2]
mg.show( locals(),
colors = {id(highlight): "red" }, # set color to red
vertical_orientations = {id(highlight): False }, # set horizontal orientation
slicers = {id(highlight): Slicer()} # set no slicing
)Different extensions are available for types from other Python packages.
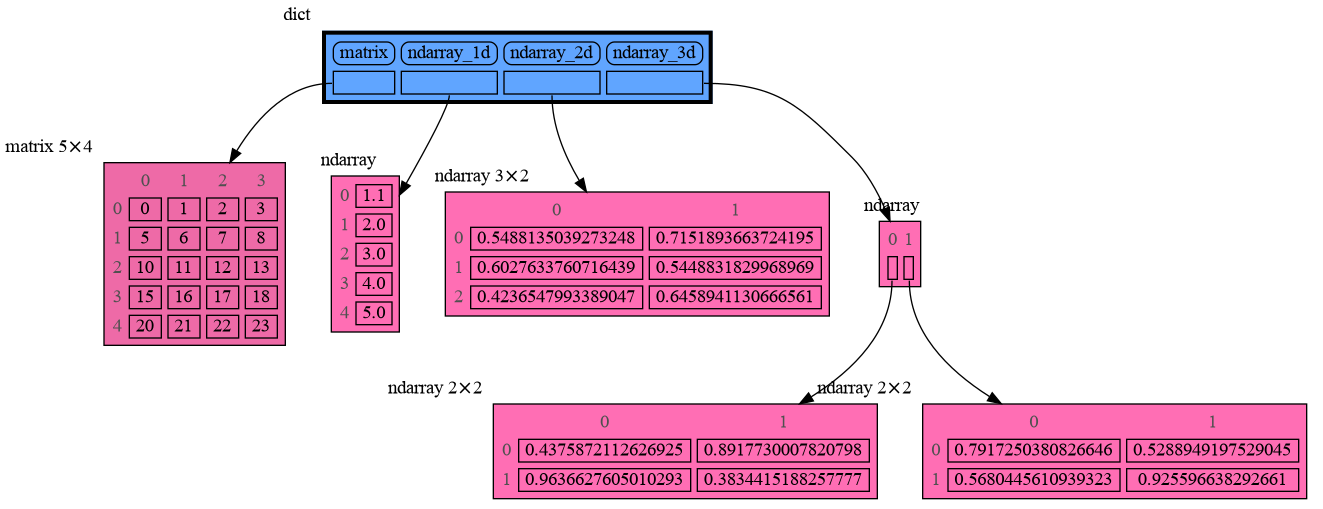
Numpy types array and matrix and ndarray can be graphed with "memory_graph.extension_numpy":
import memory_graph as mg
import numpy as np
import memory_graph.extension_numpy
np.random.seed(0) # use same random numbers each run
array = np.array([1.1, 2, 3, 4, 5])
matrix = np.matrix([[i*20+j for j in range(20)] for i in range(20)])
ndarray = np.random.rand(20,20)
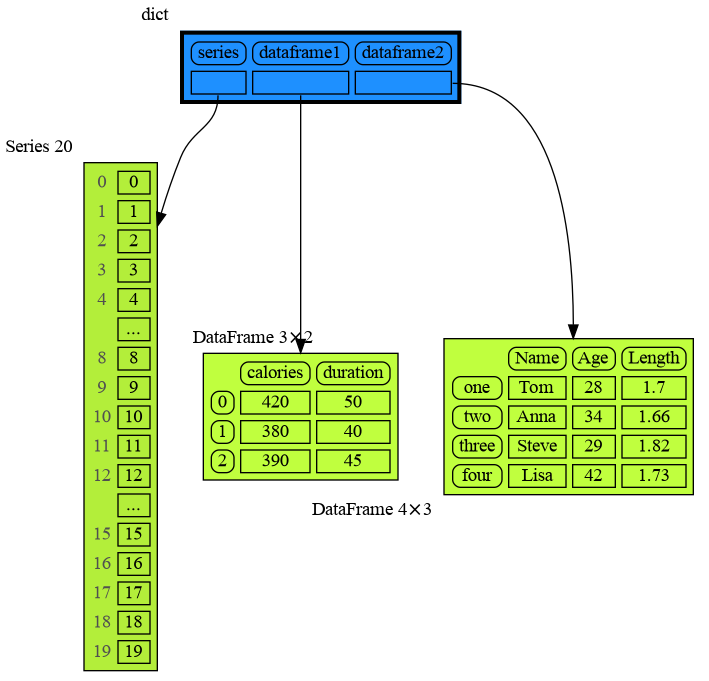
mg.show(locals())Pandas types Series and DataFrame can be graphed with "memory_graph.extension_pandas":
import memory_graph as mg
import pandas as pd
import memory_graph.extension_pandas
series = pd.Series( [i for i in range(20)] )
dataframe1 = pd.DataFrame({ "calories": [420, 380, 390],
"duration": [50, 40, 45] })
dataframe2 = pd.DataFrame({ 'Name' : [ 'Tom', 'Anna', 'Steve', 'Lisa'],
'Age' : [ 28, 34, 29, 42],
'Length' : [ 1.70, 1.66, 1.82, 1.73] },
index=['one', 'two', 'three', 'four']) # with row names
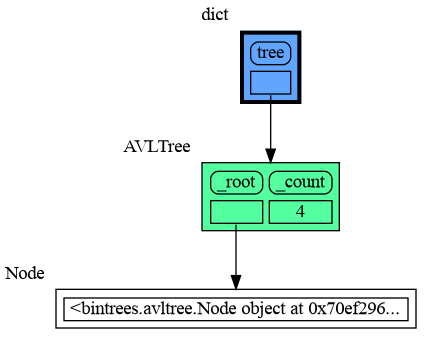
mg.show(locals())This section is likely to change. Sometimes the introspection fails or is not as desired. For example the bintrees.avltree.Node object doesn't show any attributes in the graph below.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
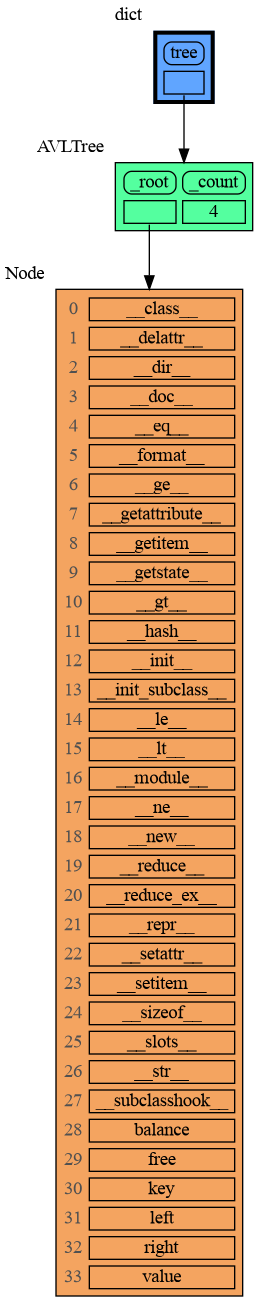
mg.show(locals())A useful start is to give it some color, show the list of all its attributes using dir(), and setting an empty Slicer to see the attribute list in full.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
mg.config.type_to_color[bintrees.avltree.Node] = "sandybrown"
mg.config.type_to_node[bintrees.avltree.Node] = lambda data: mg.node_linear.Node_Linear(data,
dir(data))
mg.config.type_to_slicer[bintrees.avltree.Node] = mg.slicer.Slicer()
mg.show(locals())Next figure out what are the attributes you want to graph and choose a Node type, there are four options:
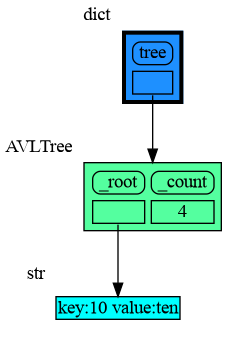
Node_base is a leaf node (with no children) and shows just a single value.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
mg.config.type_to_color[bintrees.avltree.Node] = "sandybrown"
mg.config.type_to_node[bintrees.avltree.Node] = lambda data: mg.node_base.Node_Base(f"key:{data.key} value:{data.value}")
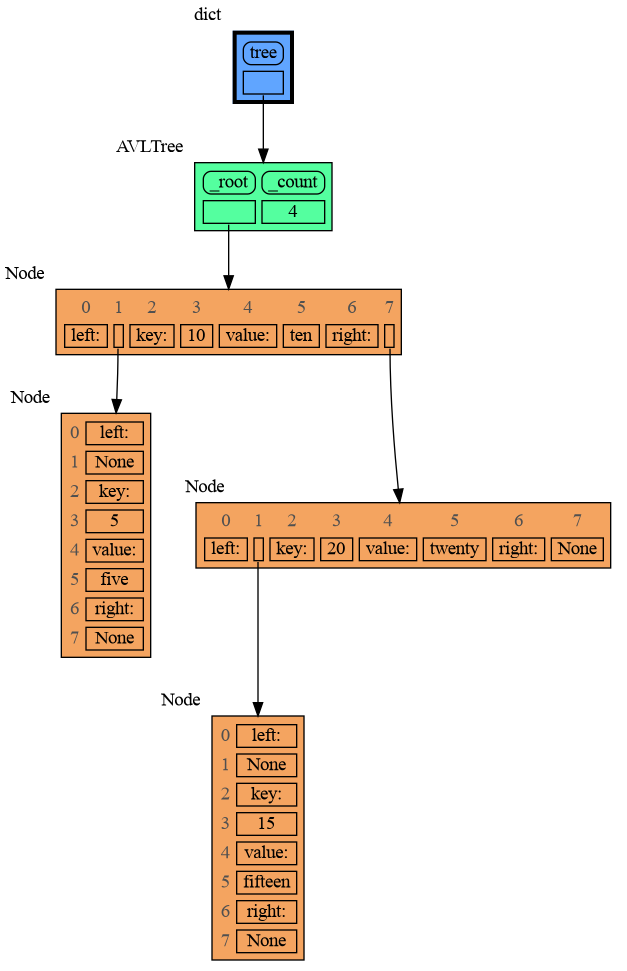
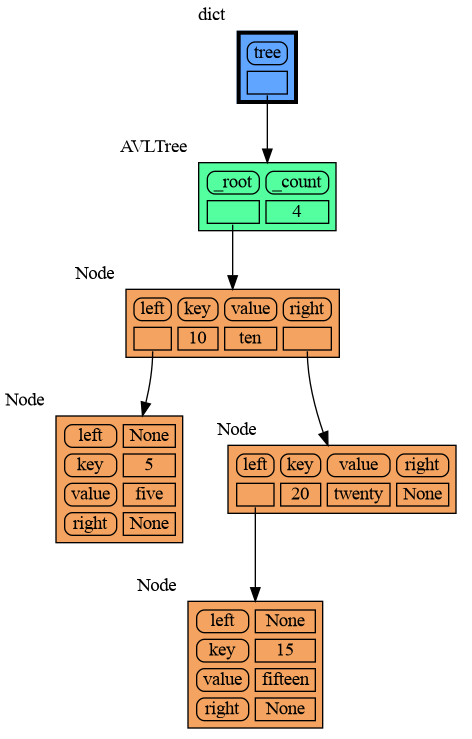
mg.show(locals())Node_Linear shows all the values in a line like a list.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
mg.config.type_to_color[bintrees.avltree.Node] = "sandybrown"
mg.config.type_to_node[bintrees.avltree.Node] = lambda data: mg.node_linear.Node_Linear(data,
['left:', data.left,
'key:', data.key,
'value:', data.value,
'right:', data.right] )
mg.show(locals())Node_Key_Value shows key-value pairs like a dictionary. Note the required items() call at the end.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
mg.config.type_to_color[bintrees.avltree.Node] = "sandybrown"
mg.config.type_to_node[bintrees.avltree.Node] = lambda data: mg.node_key_value.Node_Key_Value(data,
{'left': data.left,
'key': data.key,
'value': data.value,
'right': data.right}.items() )
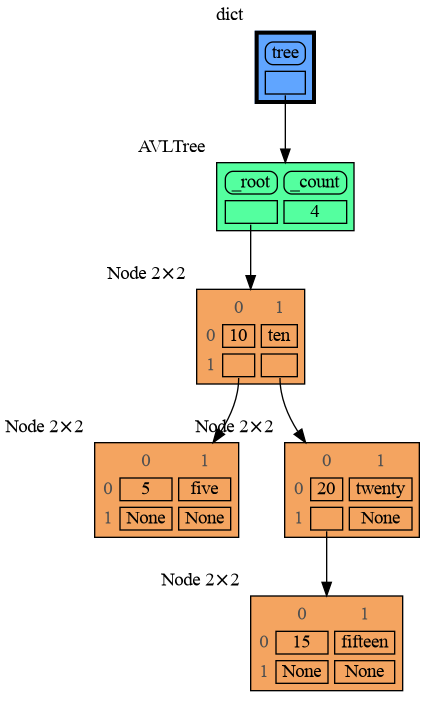
mg.show(locals())Node_Table shows all the values as a table.
import memory_graph as mg
import bintrees
# Create an AVL tree
tree = bintrees.AVLTree()
tree.insert(10, "ten")
tree.insert(5, "five")
tree.insert(20, "twenty")
tree.insert(15, "fifteen")
mg.config.type_to_color[bintrees.avltree.Node] = "sandybrown"
mg.config.type_to_node[bintrees.avltree.Node] = lambda data: mg.node_table.Node_Table(data,
[[data.key, data.value],
[data.left, data.right]] )
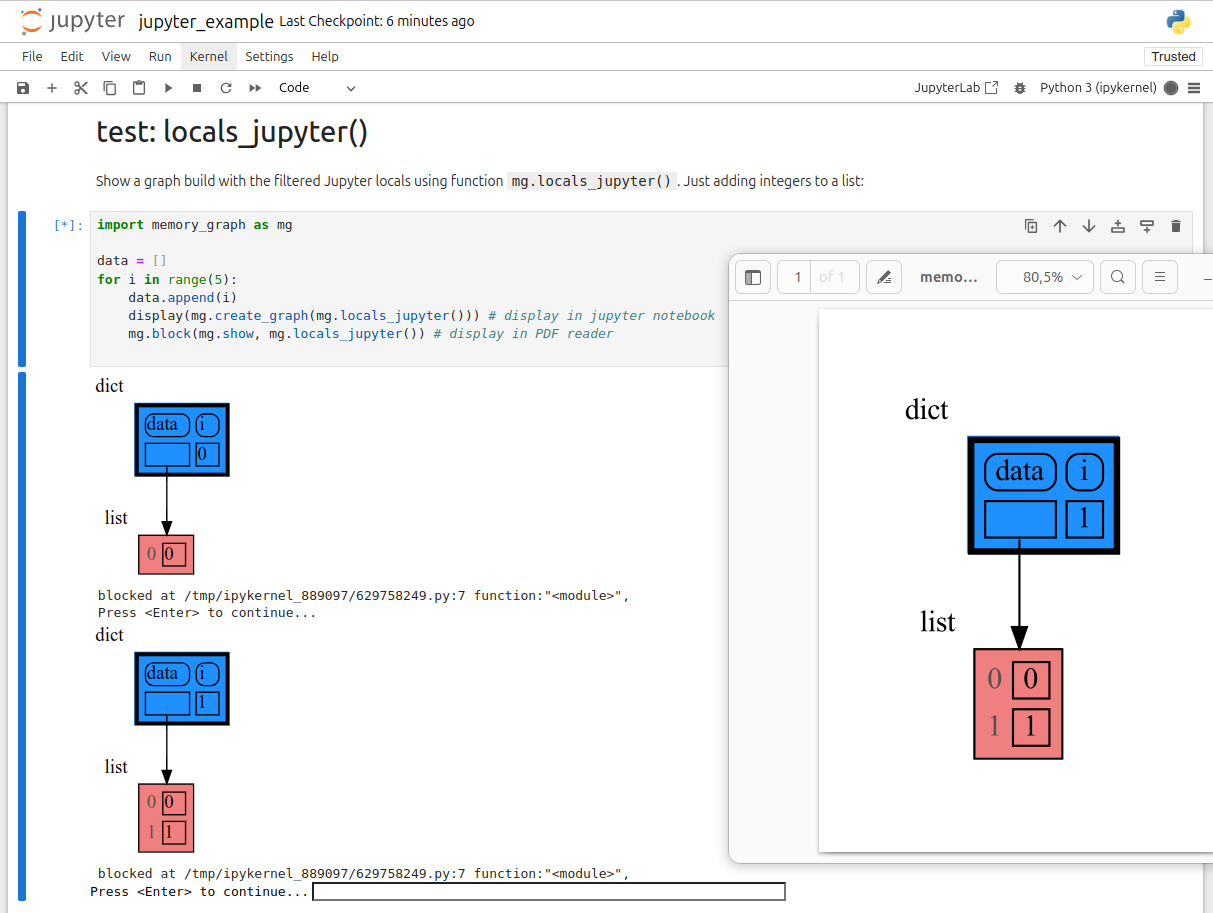
mg.show(locals())In Jupyter Notebook locals() has additional variables that cause problems in the graph, use mg.locals_jupyter() to get the local variables with these problematic variables filtered out. Use mg.get_call_stack_jupyter() to get the whole call stack with these variables filtered out.
We can use mg.show() and mg.render() in a Jupyter Notebook, but alternatively we can also use mg.create_graph() to create a graph and the display() function to render it inline with for example:
display( mg.create_graph(mg.locals_jupyter()) ) # display the local variables inline
mg.block(display, mg.create_graph(mg.locals_jupyter()) ) # the same but blockedSee for example jupyter_example.ipynb.
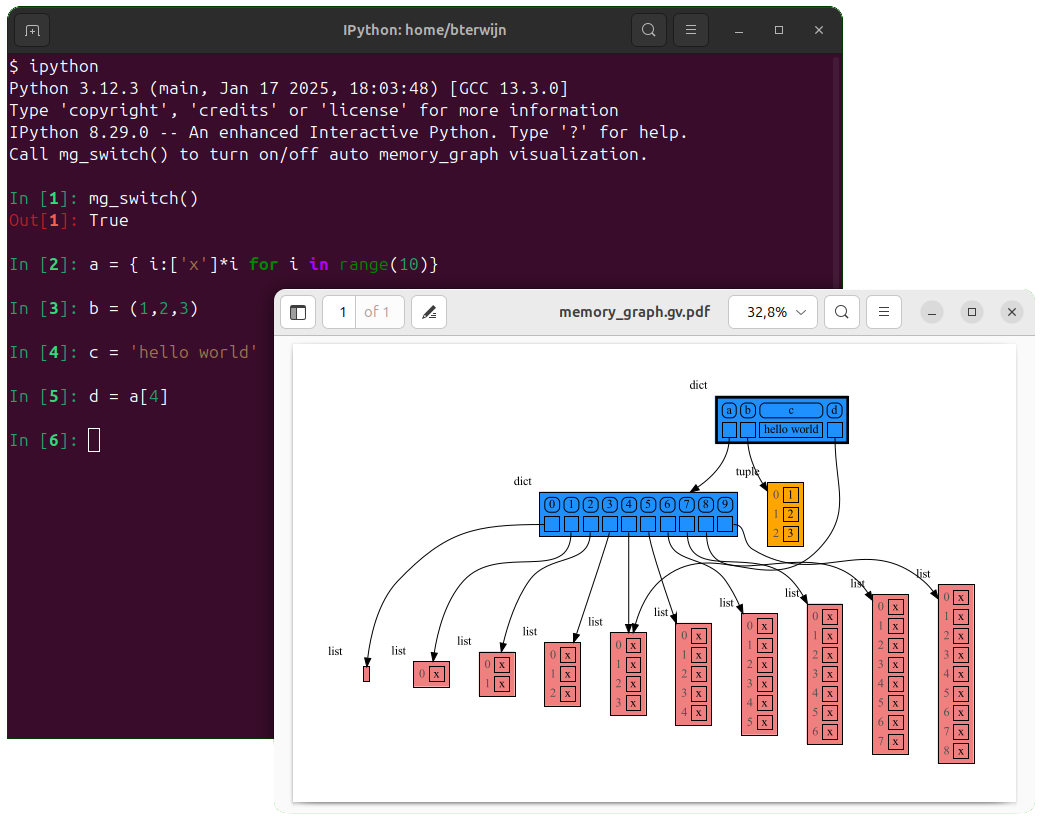
In ipython locals() has additional variables that cause problems in the graph, use mg.locals_ipython() to get the local variables with these problematic variables filtered out. Use mg.get_call_stack_ipython() to get the whole call stack with these variables filtered out.
Additionally install file auto_memory_graph.py in the ipython startup directory:
- Linux/Mac: ~/.ipython/profile_default/startup/
- Windows: %USERPROFILE%.ipython\profile_default\startup\
Then after starting 'ipython' call function mg_switch() to turn on/off the automatic visualization of local variables after each command.
We can also run memory_graph in the browser: Pyodide Example
-
Adobe Acrobat Reader doesn't refresh a PDF file when it changes on disk and blocks updates which results in an
Could not open 'somefile.pdf' for writing : Permission deniederror. One solution is to install a PDF reader that does refresh (Evince, Okular, SumatraPDF, ...) and set it as the default PDF reader. Another solution is torender()the graph to a different output format and to open it manually. -
When graph edges overlap it can be hard to distinguish them. Using an interactive graphviz viewer, such as xdot, on a '*.gv' DOT output file will help.