-
Notifications
You must be signed in to change notification settings - Fork 147
#3 Segmentation Algorithm #147
#3 Segmentation Algorithm #147
Conversation
prediction/requirements/local.txt
Outdated
| @@ -1,4 +1,4 @@ | |||
| -r base.txt | |||
| flake8==3.3.0 | |||
| pytest==3.1.3 | |||
| pylidc==0.1.8 | |||
| pylidc==0.1.9 | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This fixed a bug in to_volume, which we'll need later.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hm, I wonder if there's a better way of storing these kind of comments. Can we do
pylidc==0.1.9 # comment here
or at least put it in the commit. Putting in the PR review is really a poor place for it IMHO.
| @@ -221,9 +220,6 @@ def predict_cubes(model_path, patient_id, magnification=1, ext_name=""): # noqa | |||
| if cube_mask.sum() < 2000: | |||
| skipped_count += 1 | |||
| else: | |||
| if CROP_SIZE != CUBE_SIZE: | |||
| cube_img = helpers.rescale_patient_images2(cube_img, (CUBE_SIZE, CUBE_SIZE, CUBE_SIZE)) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The only case in which rescale_patient_images2 would be executed, which never happens (CROP_SIZE is once set to CUBE_SIZE and not modified thereafter. Thus, this branch and rescale...2 could be removed.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Again, comments here are great but...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm not completely sure how I should comment in the code base that I removed a method that hasn't been used at all?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well, if there is no need for a branch/code but someone later might think there should be that is surely worth a comment around where it "could" be...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I added a comment in 8416d58
| if scan is None: | ||
| print("Scan for path '{}' was not found".format(path)) | ||
| continue | ||
| vol = scan.to_volume(verbose=False) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
In order to get the volume of a patient scan, the leading zeros of the dicom file names had to be removed.
| def get_data_shape(): | ||
| dicom_paths = get_dicom_paths() | ||
| max_x, max_y, max_z = get_max_scaled_dimensions(dicom_paths) | ||
| return 128, 128, 128, 1 # TODO: dynamically crop shape based on maximal axes |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We need a fixed input (and thus output) size of the model. Something like taking the biggest spreads in every dimension would be possible (as it would be implemented by get_max_scaled_dimensions) but we would then be supposed to pad all images to this size. My memory was not enough for this so I decided to temporarily set the shape to a smaller, fixed one.
| return model | ||
|
|
||
|
|
||
| def unet_model_3d(input_shape, downsize_filters_factor=1, pool_size=(2, 2, 2), n_labels=1, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The U-Net uses a fully-convolutional architecture consisting of an encoder and a decoder. The encoder is able to capture contextual information while the decoder enables precise localization. Using a 3D U-Net might enable the algorithm to segment nodules better since it also gets the Z axis of the data. However, we clearly have to resize the image before passing it to the U-Net since the author states that having images of the size 144x144x144 results in 32 GB of memory consumption. We could also use 2D approaches of course, but it's questionable how we can make the algorithm aware of following and succeeding scan slices (perhaps using LSTMs?)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
These review comments are great - wouldn't we all be better served if they were in the code itself? That way they will last as this PR will eventually be somewhat difficult to find...? Just want to make sure we get good value from your effort of writing them!
707e012 to
e7321ef
Compare
|
This looks great. I think a 3D U-Net (sometimes called V-NET?) would probably take the prize for segmentation, but as you say, memory is a concern with 1mm voxel sizes. I don't think 32Gb memory use for evaluation can be right though - I'm currently training a VNET on 2 GPUS for an unrelated project, and with a batchsize of 4, I'm, still not out of memory. For inference, the memory use for a 144x144x144 volume should be less than 1Gb. I am wondering how we benchmark these approaches fairly. We can't really evaluate on LIDC, as that's what everyone will use to train the models. Do we have a good validation dataset? I only have access to radiotherapy ones, and that's not quite the same thing. |
|
Thanks @dchansen ! |
|
I'll check my exact configuration and memory as soon as I can, though it might not be before Wednesday. Sorry. I think it was 128x128x128, but I'll check. Total memory was 2x12Gb. I believe part of the efficiency of VNET comes from the use of strided convolutions, rather than pooling layers. I do know that strided convolutions can lead to checkerboard artefacts though, so it might not give as nice results. The original VNET paper used a side of 128x128x64, and was able to fit 2 batches in 8Gb of memory for training. That should be enough to fit most (all?) nodules. My concern with LIDC is that it might encourage overfitting to that dataset. Doing something like 5-fold cross validation would be quite difficult, as some of these models literally take weeks to train on a single Titan X. |
| '1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192']""" | ||
| if in_docker: | ||
| return glob.glob("../images_full/LIDC-IDRI-*/**/**") | ||
| else: |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Why the else if we have unconditional return flow on the previous line?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
If the method is not executed from within the Docker environment, the LIDC images aren't at /images_full but at test/assets/... . If you want to train the model, it would be recommended to do that outside of the Docker environment as it lacks of CUDA support. To also be able to use the GPU from within Docker, we'd be supposed to use nvidia-docker. Thus, I thought it would make sense to have a switch in the method to also get correct DICOM paths if you're outside of the Docker environment, inside of the prediction package and trying to train your model. Does that make sense?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think what @lamby is saying is that you could refactor to something like this
if in_docker:
return glob.glob("../images_full/LIDC-IDRI-*/**/**")
return glob.glob("../tests/assets/test_image_data/full/LIDC-IDRI-*/**/**")or (even better) just use one return
if in_docker:
path = glob.glob("../images_full/LIDC-IDRI-*/**/**")
else:
path = glob.glob("../tests/assets/test_image_data/full/LIDC-IDRI-*/**/**")
return pathThere was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ack
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done in 28660bf
| e.g. ['../images_full/LIDC-IDRI-0001/1.3.6.1.4.1.14519.5.2.1.6279.6001.298806137288633453246975630178/' \ | ||
| '1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192']""" | ||
| if in_docker: | ||
| return glob.glob("../images_full/LIDC-IDRI-*/**/**") |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
We should always be using os.path.join :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ping on this one? :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done in 28660bf
| axis: index of the axis that should be changeable using the slider | ||
| is_mask: whether the cuboid that should be plotting is a binary mask | ||
| """ | ||
| import matplotlib.pyplot as plt |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
How come the inline imports? (please explain in a comment, etc.)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It's unnecessary, thanks ;)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Done in a1d18ab
| INTERMEDIATE_MALICIOUS = 3 | ||
|
|
||
| current_dir = os.path.dirname(os.path.realpath(__file__)) | ||
| assets_dir = os.path.abspath(os.path.join(current_dir, '../assets')) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
how about something like this?
...
parent_dir = os.path.dirname(current_dir)
assets_dir = os.path.abspath(os.path.join(parent_dir, 'assets'))There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
there's also 2 other times this same code block is used... you could consider making it a top level constant.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
added get_assets_dir() in 28660bf
|
|
||
| for index, path in enumerate(dicom_paths[batch_index:batch_index + CUBOID_BATCH]): | ||
| lidc_id = path.split('/')[5] | ||
| patient_id = path.split('/')[-1] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
minor nitpick, but how about setting the result of path.split('/') to a variable so that the split method is only called once?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Also, take a look at os.path.splitext, os.path.split, or PurePath.parts which should be more platform independent than split('/'). If none of those work, use os.path.sep instead of hardcoding /.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks, implemented in 28660bf
| dicom_paths = get_dicom_paths() | ||
| max_x, max_y, max_z = get_max_scaled_dimensions(dicom_paths) | ||
| return 128, 128, 128, 1 # TODO: dynamically crop shape based on maximal axes | ||
| return max_x, max_y, max_z, 1 |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
It's still a bit odd having two return statements. If you want to convey the same message, I'd suggest you move the first return statement to the end, and then comment out the other parts of the function. Also, could you add your above comment to the code itself so that others know why it's been commented out?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You're right, I commented the method properly now :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'd suggest you move the first return statement to the end, and then comment out the other parts of the function.
Leaving commented-out code in PRs doesn't seem like a good practice. If there's a piece that is broken or semi-working but still merits being merged, perhaps the problems and TODOs can be tracked as issues instead.
A # TODO: [...] comment used sparingly may be okay _ to explain to readers_ why something it the way it is -- it's not a helpful way to track outstanding work.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
So I should add an issue for that?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
prediction/requirements/local.txt
Outdated
| @@ -1,4 +1,4 @@ | |||
| -r base.txt | |||
| flake8==3.3.0 | |||
| pytest==3.1.3 | |||
| pylidc==0.1.8 | |||
| pylidc==0.1.9 | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hm, I wonder if there's a better way of storing these kind of comments. Can we do
pylidc==0.1.9 # comment here
or at least put it in the commit. Putting in the PR review is really a poor place for it IMHO.
| @@ -221,9 +220,6 @@ def predict_cubes(model_path, patient_id, magnification=1, ext_name=""): # noqa | |||
| if cube_mask.sum() < 2000: | |||
| skipped_count += 1 | |||
| else: | |||
| if CROP_SIZE != CUBE_SIZE: | |||
| cube_img = helpers.rescale_patient_images2(cube_img, (CUBE_SIZE, CUBE_SIZE, CUBE_SIZE)) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Again, comments here are great but...
| e.g. ['../images_full/LIDC-IDRI-0001/1.3.6.1.4.1.14519.5.2.1.6279.6001.298806137288633453246975630178/' \ | ||
| '1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192']""" | ||
| if in_docker: | ||
| return glob.glob("../images_full/LIDC-IDRI-*/**/**") |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ping on this one? :)
| '1.3.6.1.4.1.14519.5.2.1.6279.6001.179049373636438705059720603192']""" | ||
| if in_docker: | ||
| return glob.glob("../images_full/LIDC-IDRI-*/**/**") | ||
| else: |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ack
|
@lamby I already updated the if CROP_SIZE != CUBE_SIZE:
cube_img = helpers.rescale_patient_images2stuff need comments? If we're honest, this shouldn't even have made it to the current code base - I introduced it in #118 but only noticed now that it currently isn't executed and was probably used by Julian de Wit to play around with the |
|
By "comments" I mean that — in various places in this PR — you have added extremely helpful descriptive comments on the changes, but they should live in the code otherwise your input of time and energy will very likely be "lost" in the depths of GitHub :( |
0fe0893 to
25ba711
Compare
| in_docker: whether this method is invoked from within docker or from the prediction directory | ||
| """ | ||
| if in_docker: | ||
| paths = glob.glob(os.path.join('..', 'images_full', 'LIDC-IDRI-*', '**', '**')) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hm, relative paths? Smells wrong...
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
fixed in 8416d58
| @@ -221,9 +220,6 @@ def predict_cubes(model_path, patient_id, magnification=1, ext_name=""): # noqa | |||
| if cube_mask.sum() < 2000: | |||
| skipped_count += 1 | |||
| else: | |||
| if CROP_SIZE != CUBE_SIZE: | |||
| cube_img = helpers.rescale_patient_images2(cube_img, (CUBE_SIZE, CUBE_SIZE, CUBE_SIZE)) | |||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Well, if there is no need for a branch/code but someone later might think there should be that is surely worth a comment around where it "could" be...
| input_data = padded_data | ||
|
|
||
| output_data = model.predict(input_data) | ||
| segment_path = os.path.join(os.path.dirname(__file__), 'assets', "lung-mask.npy") |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Would prefer more constants than the use of underscoreunderscorefileunderscoreunderscore :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
fixed in 8416d58
|
@lamby Would you mind reviewing again? |
|
|
||
|
|
||
| def get_assets_dir(): | ||
| return os.path.abspath(os.path.join(os.getcwd(), 'src', 'algorithms', 'segment', 'assets')) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is surely just the same as underscore underscore file underscore underscore ?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I also see some other uses of ../images still lingering around? Not sure if an oversight, or...?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do the constants introduced in 773df14 make sense?
| in_docker: whether this method is invoked from within docker or from the prediction directory | ||
| """ | ||
| if in_docker: | ||
| paths = glob.glob(os.path.join('/images_full', 'LIDC-IDRI-*', '**', '**')) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is /images_full correct here? Looks like an absolute path. Again, we should be using some kind of constant, not hardcoding these deep under prediction/src/algorithms/segment/src/data_generation.py :)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
In the docker container, the full LIDC images are mounted under /images_full/... . test_correct_paths should ensure that the folders exist. However, you're right - I'll add constants to our settings - or should I define them somewhere else?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Like it,except for the
prediction/config.py
Outdated
| SEGMENT_ASSETS_DIR = os.path.abspath(os.path.join(os.getcwd(), 'src', 'algorithms', 'segment', 'assets')) | ||
| DICOM_PATHS_DOCKER = glob.glob(os.path.join('/images_full', 'LIDC-IDRI-*', '**', '**')) | ||
| DICOM_PATHS_LOCAL = glob.glob( | ||
| os.path.join(os.getcwd(), 'src', 'tests', 'assets', 'test_image_data', 'full', 'LIDC-IDRI-*', '**', '**')) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Hm, do we want to glob.glob on module import? Doesn't feel right.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Does using constant wildcards as introduced in cd7d39a make more sense?
|
LGTM. @reubano ? |
prediction/config.py
Outdated
| DICOM_PATHS_LOCAL = glob.glob( | ||
| os.path.join(os.getcwd(), 'src', 'tests', 'assets', 'test_image_data', 'full', 'LIDC-IDRI-*', '**', '**')) | ||
| DICOM_PATHS_DOCKER_WILDCARD = os.path.join('/images_full', 'LIDC-IDRI-*', '**', '**') | ||
| DICOM_PATHS_LOCAL_WILDCARD = os.path.join(os.getcwd(), 'src', 'tests', 'assets', 'test_image_data', 'full', |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'd juuust add a brief comment along the lines of "these are expanded at runtime" so that nobody gets too confused.
(Also I don't quite like the use of getcwd - is that "fixed"? Seems more sensible to use underscoreunderscorefileunderscoreunderscore)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You have to be in the prediction directory otherwise none of your code imports for the prediction code would work since this directory is copied into the prediction docker container. However, I can use __file__ here as well.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
You have to be in the prediction directory
Lets fix that another time, but let's at least not make the situation "worse" ;)
|
Hi, |
|
Hi @rracinskij , |
|
Thank you, @WGierke. Another newbie question - is there a training dataset with segmentation masks available or should the masks be derived from the annotations first? |
|
@rracinskij We use the scans of the LIDC dataset. You can already find 3 patient scans in tests/assets/test_image_data/full. The neat thing is that there is a library (pylidc) that makes it really easy to query scan and annotation information. After downloading the dataset, set up pylidc by creating a |
|
@WGierke Sounds reasonable :) |
|
LGTM (as before). @reubano ? |
|
It's looking good! With a few tweaks I got everything to run and was able to check the output of the routes via Even though #142 won't be merged, I did like the simplicity of it since it doesn't seem to require training a model. I also like that it includes a So my question to the both of you is this: Is there value in porting the Also, one thing I wasn't able to resolve was the inability to directly compare both algorithms. This one errors when passed |
|
aforementioned tweaks prediction/src/algorithms/identify/prediction.py | 3 ++-
prediction/src/algorithms/segment/src/training.py | 8 ++++++--
prediction/src/preprocess/lung_segmentation.py | 2 --# diff --git a/prediction/src/algorithms/identify/prediction.py b/prediction/src/algorithms/identify/prediction.py
# --- a/prediction/src/algorithms/identify/prediction.py
# +++ b/prediction/src/algorithms/identify/prediction.py
from keras.layers import Input, Convolution3D, MaxPooling3D, Flatten, AveragePoo
from keras.metrics import binary_accuracy, binary_crossentropy, mean_absolute_error
from keras.models import Model
from keras.optimizers import SGD
-from src.preprocess.lung_segmentation import rescale_patient_images
+
+from ...preprocess.lung_segmentation import rescale_patient_images
CUBE_SIZE = 32
MEAN_PIXEL_VALUE = 41# diff --git a/prediction/src/algorithms/segment/src/training.py b/prediction/src/algorithms/segment/src/training.py
# --- a/prediction/src/algorithms/segment/src/training.py
# +++ b/prediction/src/algorithms/segment/src/training.py
import os
import numpy as np
import pylidc as pl
-from config import Config
from keras.callbacks import ModelCheckpoint
from keras.models import load_model
-from src.preprocess.lung_segmentation import save_lung_segments
from .model import simple_model_3d
+from ....preprocess.lung_segmentation import save_lung_segments
+
+try:
+ from .....config import Config
+except ValueError:
+ from config import Config
def get_data_shape():# diff --git a/prediction/src/preprocess/lung_segmentation.py b/prediction/src/preprocess/lung_segmentation.py
# --- a/prediction/src/preprocess/lung_segmentation.py
# +++ b/prediction/src/preprocess/lung_segmentation.py
from skimage.measure import label, regionprops
from skimage.morphology import disk, binary_erosion, binary_closing
from skimage.segmentation import clear_border
-from ..algorithms.identify.helpers import rescale_patient_images |
|
|
|
@WGierke This should make merging a bit easier... after downloading patch.txt just perform the following: # assuming you are on commit #9123a2
git apply --check patch.txt
git apply patch.txt
git commit -m 'Fix conflicts and bugs' # or whatever other message you preferedit: updated patch file |
|
Thanks a lot @reubano . Much appreciated :) |
|
Minor error on travis. just hold tight... |
No prob. And here's a new patch file which should pass travis... |
|
Thanks @reubano - works like a charm :) |
|
Awesome! Can you squash to 1 commit? |
8d412a0 to
0445e5b
Compare
0445e5b to
9a42000
Compare
|
@reubano Done. Travis had some temporary issues so I had to |
|
As demanded by the ticket, I implemented a very simple segmentation algorithm that can be trained using Keras, its model is saved in the
assetsfolder and that can predict nodules. The model is still pretty shitty, so hopefully there are some more experiences data scientists than me around to improve it :) However, the "environment" to train the model should be implemented by this PR.Description
I created training data pairs based on the LIDC dataset using pylidc. The label image is a binary mask whose pixels are 1 if there is an annotation in the LIDC dataset whose malice is bigger or equal to 3 (out of 5). The label images are saved in the
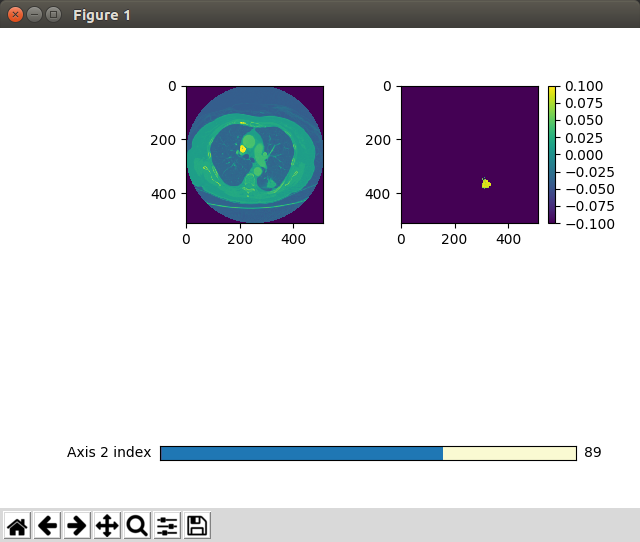
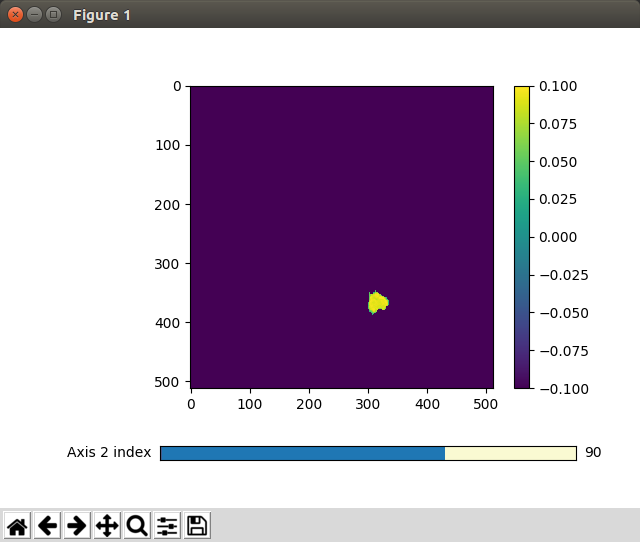
prediction/src/algorithms/segment/assets/directory. Since some 3D arrays are hard to imagine, I added two methods that help to visualize these arrays by navigating through one dimension and simultaneously displaying the remaining two ones:display_training_pair()shows the input image and the binary output mask. Note the segmented annotation on the right hand side and the slider for the Z axis at the bottom.When the mask is being applied on the input data,
cuboid_show_slidershows us:prepare_training_data() in prediction/src/algorithms/segment/src/data_generation.py saves a npy file including the binary segmented mask of a lung for each patient.
train() in prediction/src/algorithms/segment/src/training.py trains the model based on the previously generated binary masks and the rescaled volume images of each DICOM scan. The model configuration is saved in prediction/src/algorithms/segment/src/model.py.
Reference to official issue
This addresses #3
Motivation and Context
We want to suggest the radiologist to have a look at nodules that are automatically found by an algorithm of us.
How Has This Been Tested?
I wrote a test that checks whether the response of the segmentation endpoint has the correct format.
CLA
I'd highly appreciate any feedback!