-
Notifications
You must be signed in to change notification settings - Fork 302
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Precomputed Multiresolution mesh generation issue #272
Comments
|
If you adjust the "Resolution (mesh)" control under the "Render" tab, you can see that the finest resolution mesh (lod0) is displayed incorrectly ("exploded" relative to the other levels of detail), but the other two levels of detail appear to be displayed correctly. As for why that is happening, I do not know. I haven't attempted to examine the mesh data directly. However, if you share the code you are using to generate it, I can take a look and see what the issue might be. |
|
Yes indeed it is the finest resolution mesh, that is what I am reporting. Here are the relevant parts of the code generating the mesh: class Quantize():
def __init__(self, fragment_origin, fragment_shape, input_origin, quantization_bits):
self.upper_bound = np.iinfo(np.uint32).max >> (np.dtype(np.uint32).itemsize*8 - quantization_bits)
self.scale = self.upper_bound / fragment_shape
self.offset = input_origin - fragment_origin + 0.5/self.scale
def __call__(self, v_pos):
output = np.minimum(self.upper_bound, np.maximum(0, self.scale*(v_pos + self.offset))).astype(np.uint32)
return output
def cmp_zorder(lhs, rhs) -> bool:
def less_msb(x: int, y: int) -> bool:
return x < y and x < (x ^ y)
# Assume lhs and rhs array-like objects of indices.
assert len(lhs) == len(rhs)
# Will contain the most significant dimension.
msd = 2
# Loop over the other dimensions.
for dim in [1, 0]:
# Check if the current dimension is more significant
# by comparing the most significant bits.
if less_msb(lhs[msd] ^ rhs[msd], lhs[dim] ^ rhs[dim]):
msd = dim
return lhs[msd] - rhs[msd]
def generate_mesh_decomposition(verts, faces, nodes_per_dim, bits):
# Scale our coordinates.
scale = nodes_per_dim/(verts.max(axis=0) - verts.min(axis=0))
verts_scaled = scale*(verts - verts.min(axis=0))
# Define plane normals and create a trimesh object.
nyz, nxz, nxy = np.eye(3)
mesh = trimesh.Trimesh(vertices=verts_scaled, faces=faces)
# create submeshes.
submeshes = []
nodes = []
for x in range(0, nodes_per_dim):
mesh_x = trimesh.intersections.slice_mesh_plane(mesh, plane_normal=nyz, plane_origin=nyz*x)
mesh_x = trimesh.intersections.slice_mesh_plane(mesh_x, plane_normal=-nyz, plane_origin=nyz*(x+1))
for y in range(0, nodes_per_dim):
mesh_y = trimesh.intersections.slice_mesh_plane(mesh_x, plane_normal=nxz, plane_origin=nxz*y)
mesh_y = trimesh.intersections.slice_mesh_plane(mesh_y, plane_normal=-nxz, plane_origin=nxz*(y+1))
for z in range(0, nodes_per_dim):
mesh_z = trimesh.intersections.slice_mesh_plane(mesh_y, plane_normal=nxy, plane_origin=nxy*z)
mesh_z = trimesh.intersections.slice_mesh_plane(mesh_z, plane_normal=-nxy, plane_origin=nxy*(z+1))
# Initialize Quantizer.
quantize = Quantize(
fragment_origin=np.array([x, y, z]),
fragment_shape=np.array([1, 1, 1]),
input_origin=np.array([0,0,0]),
quantization_bits=bits
)
if len(mesh_z.vertices) > 0:
mesh_z.vertices = quantize(mesh_z.vertices)
submeshes.append(mesh_z)
nodes.append([x,y,z])
# Sort in Z-curve order
submeshes, nodes = zip(*sorted(zip(submeshes, nodes), key=cmp_to_key(lambda x, y: cmp_zorder(x[1], y[1]))))
return nodes, submeshesI call the main workhorse function quantization_bits = 16
lods = np.array([0, 1, 2])
chunk_shape = (verts.max(axis=0) - verts.min(axis=0))/2**lods.max()
grid_origin = verts.min(axis=0)
lod_scales = np.array([2**lod for lod in lods])
num_lods = len(lod_scales)
vertex_offsets = np.array([[0.,0.,0.] for _ in range(num_lods)])
fragment_offsets = []
fragment_positions = []
with open('../precompute/multi_res/mesh/1', 'wb') as f:
for scale in lod_scales[::-1]:
lod_offsets = []
nodes, submeshes = generate_mesh_decomposition(verts.copy(), faces.copy(), scale, quantization_bits)
for mesh in submeshes:
draco = trimesh.exchange.ply.export_draco(mesh, bits=16)
f.write(draco)
lod_offsets.append(len(draco))
fragment_positions.append(np.array(nodes))
fragment_offsets.append(np.array(lod_offsets))
num_fragments_per_lod = np.array([len(nodes) for nodes in fragment_positions])Here is my code writing out the info and manifest files: with open('../precompute/multi_res/mesh/1.index', 'wb') as f:
f.write(chunk_shape.astype('<f').tobytes())
f.write(grid_origin.astype('<f').tobytes())
f.write(struct.pack('<I', num_lods))
f.write(lod_scales.astype('<f').tobytes())
f.write(vertex_offsets.astype('<f').tobytes(order='C'))
f.write(num_fragments_per_lod.astype('<I').tobytes())
for frag_pos, frag_offset in zip(fragment_positions, fragment_offsets):
f.write(frag_pos.T.astype('<I').tobytes(order='C'))
f.write(frag_offset.astype('<I').tobytes(order='C'))
with open('../precompute/multi_res/mesh/info', 'w') as f:
info = {
'@type': 'neuroglancer_multilod_draco',
'vertex_quantization_bits': 16,
'transform': [1,0,0,0,0,1,0,0,0,0,1,0],
'lod_scale_multiplier': 1
}
json.dump(info, f)Thanks again |
|
Just following up on this. Any idea what might be going on, @jbms? Thanks! |
|
It seems that the issue was that the draco_encoder executable was creating a bounding box around the mesh and not respecting offsets for each octree node subvolume. Using the Draco API solved this issue. Thanks! |
|
Hi @hsidky, I'm finding your code very helpful! I was wondering what the Draco fix was to respect the bounding box. |
|
My attempt, including a hack for Draco: https://github.com/davidackerman/multiresolutionMeshes. |
Hi,
I have been working on writing code to generate precomputed multi-resolution meshes for neuroglancer and have run into a weird issue. My code seems to work fine for a single lod (1 octree node) and two lods (8 octree nodes). However, when I go to the third lod (64 octree nodes) the fragments do not piece together properly. I am working with a perfect sphere which I generated to serve as a MWE. I have also checked the discussion in #266 to make sure I haven't missed anything.
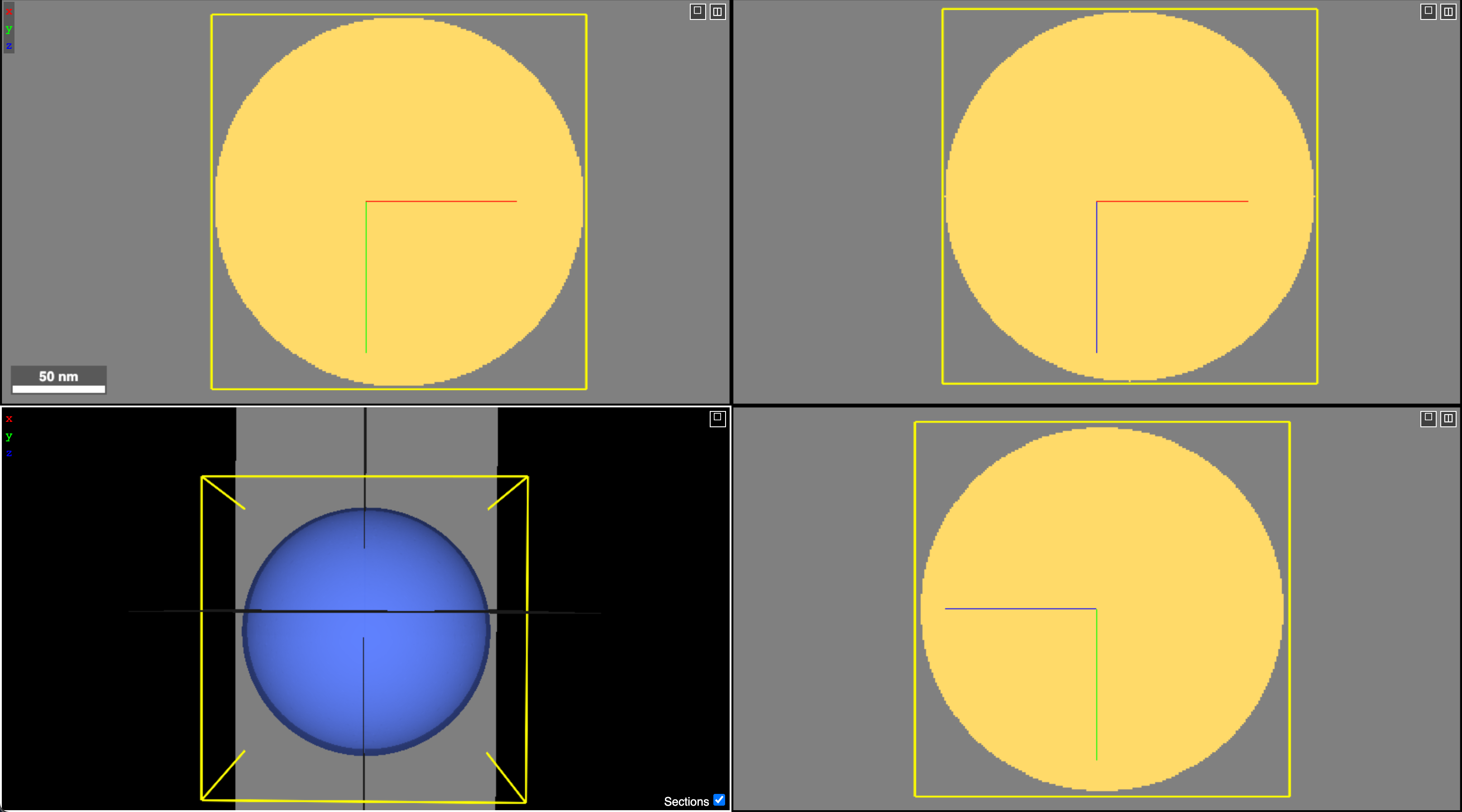
Below is a screenshot for the working LODs (they are identical so I am only sharing a single screenshot)
Here is what it looks like with 64 nodes:
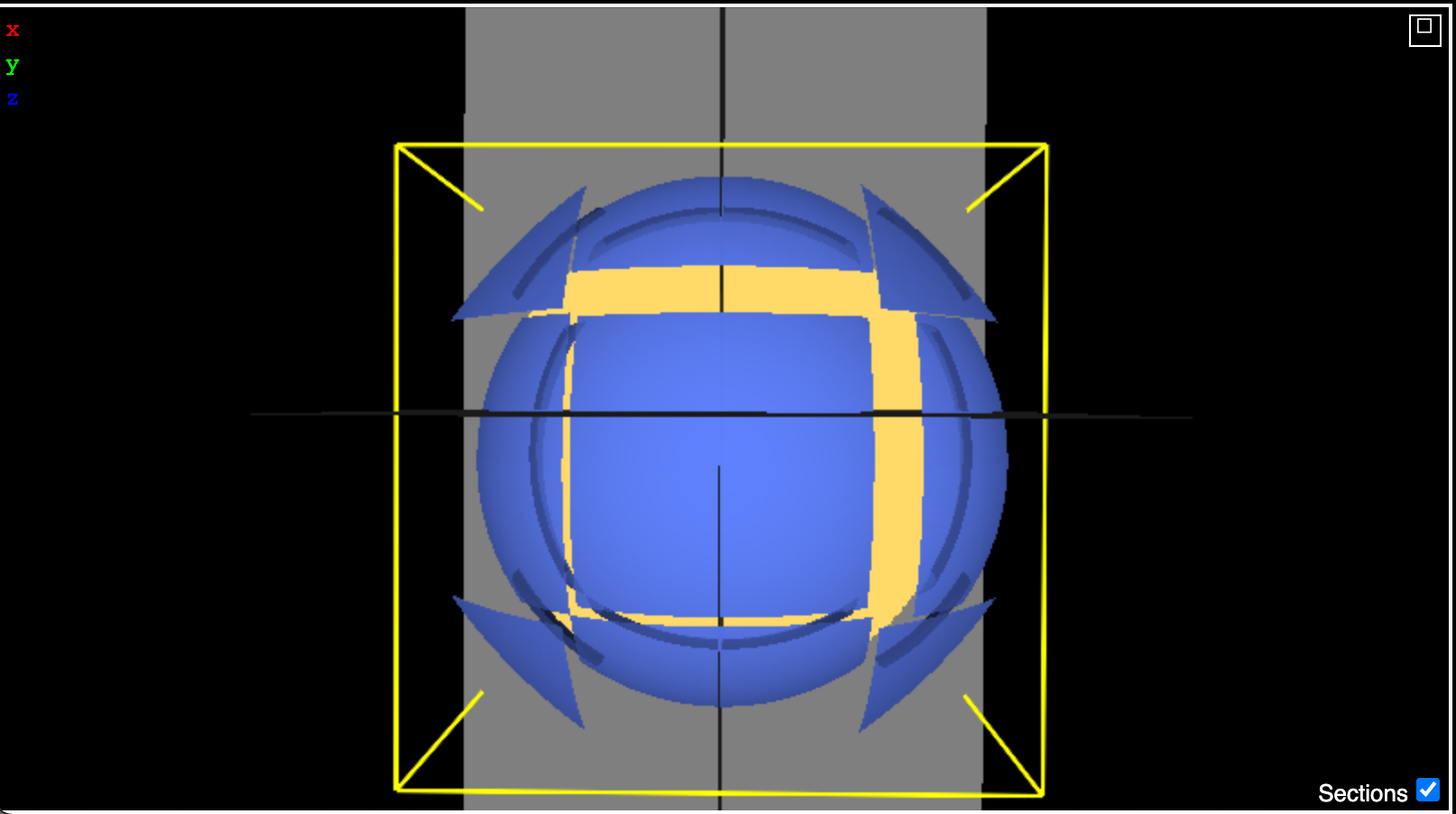
What's strange is that the patches which are offset are not necessarily single fragments. I have reconstructed the mesh using in Python from the same data I am writing to disk, and it works fine. See the image below:
I have colored every 8th fragment the same and it reproduces the decomposition pattern shown in neuroglancer. I am wondering if somehow it is related to the z-curve order but things work fine for 8 nodes so I'm not sure that's the issue. A difference between 8 nodes and 64 nodes is that all nodes contain a submesh for 8 nodes. However, for 64 nodes there are 8 empty nodes for a sphere. I have tried both excluding those and also including them with a fragment offset of 0, and the result is identical.
I am hoping someone might have an intuition for what's going on. I have attached a zip file containing my mesh folder in case that might be useful. I am also happy to provide anything else if it'll help resolve this issue.
Thanks!
The text was updated successfully, but these errors were encountered: