-
Notifications
You must be signed in to change notification settings - Fork 7
Differential Splicing
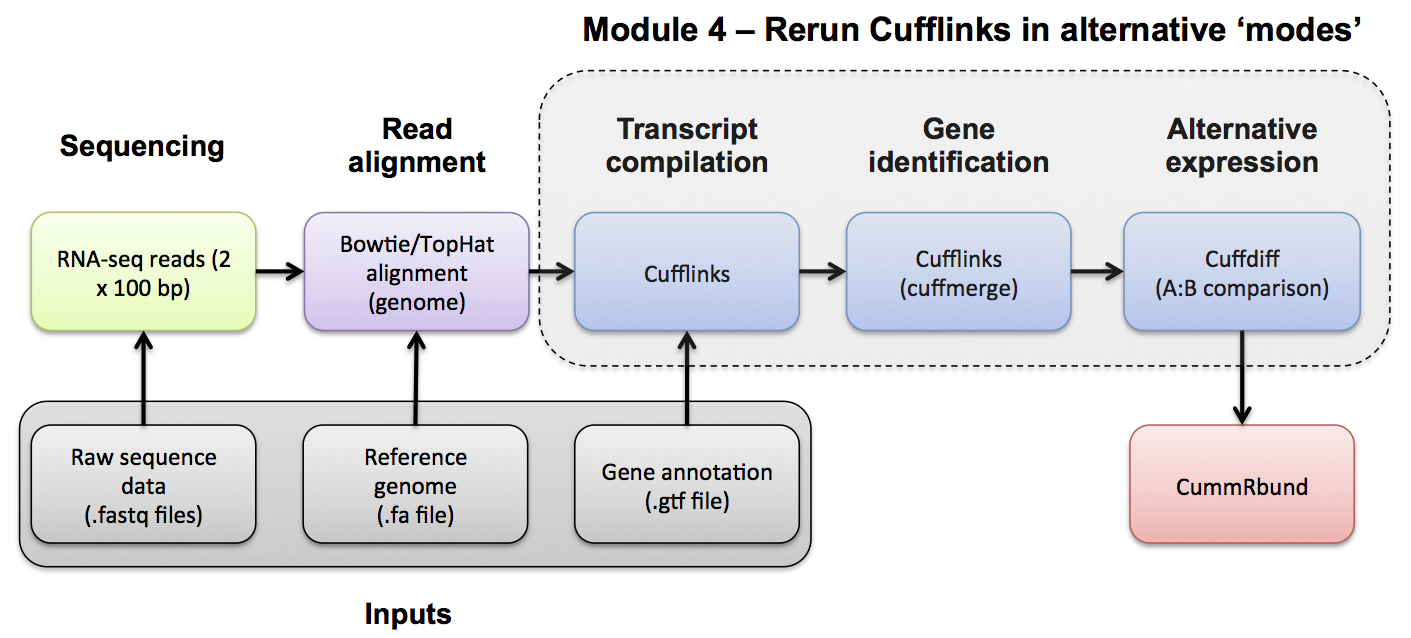
#4-iv. Differential Splicing
Use Cuffdiff to compare the UHR and HBR conditions.
Refer to the Cufflinks manual for a more detailed explanation: http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html
Cuffdiff basic usage:
cuffdiff [options] <transcripts.gtf> <sample1_hits.sam> <sample2_hits.sam> [... sampleN_hits.sam]
- Supply replicate SAMs as comma separated lists for each condition: sample1_rep1.sam,sample1_rep2.sam,...sample1_repM.sam
- '-p 8' tells cuffdiff to use eight CPUs
- '-L' tells cuffdiff the labels to use for samples
Perform UHR vs. HBR comparison, for known/novel (reference guided mode) transcripts:
cd $RNA_HOME/
mkdir -p de/tophat_cufflinks/ref_guided/
cd $RNA_HOME/alignments/tophat/
cuffdiff -p 8 -L UHR,HBR -o $RNA_HOME/de/tophat_cufflinks/ref_guided/ --frag-len-mean 262 --frag-len-std-dev 80 --no-update-check $RNA_HOME/expression/tophat_cufflinks/ref_guided/merged/merged.gtf UHR_Rep1_ERCC-Mix1/accepted_hits.bam,UHR_Rep2_ERCC-Mix1/accepted_hits.bam,UHR_Rep3_ERCC-Mix1/accepted_hits.bam HBR_Rep1_ERCC-Mix2/accepted_hits.bam,HBR_Rep2_ERCC-Mix2/accepted_hits.bam,HBR_Rep3_ERCC-Mix2/accepted_hits.bam
Perform UHR vs. HBR comparison, for known/novel (de novo mode) transcripts:
cd $RNA_HOME/
mkdir -p de/tophat_cufflinks/de_novo/
cd $RNA_HOME/alignments/tophat/
cuffdiff -p 8 -L UHR,HBR -o $RNA_HOME/de/tophat_cufflinks/de_novo/ --frag-len-mean 262 --frag-len-std-dev 80 --no-update-check $RNA_HOME/expression/tophat_cufflinks/de_novo/merged/merged.gtf UHR_Rep1_ERCC-Mix1/accepted_hits.bam,UHR_Rep2_ERCC-Mix1/accepted_hits.bam,UHR_Rep3_ERCC-Mix1/accepted_hits.bam HBR_Rep1_ERCC-Mix2/accepted_hits.bam,HBR_Rep2_ERCC-Mix2/accepted_hits.bam,HBR_Rep3_ERCC-Mix2/accepted_hits.bam
| Previous Section | This Section | Next Section | |:----------------------------------------------:|:-----------------------------------------------:|:-----------------------------------------------------------------:| | Merging | Differential Splicing | Splicing Visualization |
##Note: The current version of this tutorial is now at www.rnaseq.wiki
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
Module 4: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 5: Kallisto
Appendix: Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup