-
Notifications
You must be signed in to change notification settings - Fork 96
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Performance of different polygonizing methods analyzed #13
Comments
|
Thanks -- helpful!! |
|
@mdsumner |
|
@mdsumner but note that |
|
Looks like a candidate (or two?) to be added to |
|
Indeed, it would be awesome. Two candidates, I would say. The integer one is very useful for categorical variables, the float version is a lot slower. It's easy enough to modify |
|
GDALPolygonize is now the default in |
|
Two examples here. |
|
This really should be a huge improvement to my workflow. I'll do some test against |
|
It currently writes to disk before polygonizing; I was contemplating writing to a memory driver instead, but haven't sorted out yet how. Not sure if it would matter either. |
|
You could just use |
|
Yeah, but operating systems also use memory to buffer/cache files, so that should not be needed (I hope). |
|
Yep, was suggesting just a temporary workaround for testing purposes. |
|
I thought about squeezing this into Haven't tried out the options argument, might as well work. |
|
I'm getting a: Error in gdal_polygonize(x, mask, use_integer = use_integer, geotransform = get_geotransform(x)) :
could not find function "gdal_polygonize" |
|
Reinstall sf, using |
|
The new implementation is 2x faster than EDIT: this is on an oldish i7 machine with only rotational drives. |
|
Thanks. Yes, I chose to make this only work for a single layer, because cells are merged and this would happen differently for every layer: the input of GDALPolygonize is a single raster band. |
|
Yes, I agree with this. Should output a warning tho. |
|
Also a warning for the fact that it only polygonizes the first variable, other than the first 'layer', would be necessary. |
|
I did another test on a different dataset. The ~200x improvement! The slow part is This is on a modern laptop with a decent SSD. |
|
Is this point-interpretation (like contour,filled.contour) or area (like rasterToPolygons, spex::polygonize)? Can that be controlled with GDAL? I will check it out but keen to discuss a bit. |
|
Description here, the only controllable option I can see is
You'd set it by passing |
|
Yes, thanks! I just did some quick checks with gdal_polygonize.py - it's definitely a different thing. FWIW, I have explored the code under filled.contour and would like to bring it out for the point-interpretation, I often need it and there are ways to achieve it currently but it's pretty painful. Unioning the data structures from filled.contour with sf is probably promising (it's not exactly related to this issue here, but I wanted to air it to make sure). It's also the difference between I'm impressed that it's so fast, |
|
I agree, it would be extremely cool if we could get contour polygons out, in addition to those following the pixel boundaries. Since |
|
@mdsumner |
|
It seems that GDALContourGenerateEx has an option to create contour polygons, rather than polygon lines. Worth exploring? |
|
oh gosh - I've been scratching around - thought to search GDAL for marching squares while I was marching around but didn't actually do it! I've learnt enough about the basic problem, this looks great! |
|
Oh, it's brand new - looks like -p only since 2.4 https://gdal.org/gdal_contour.html |
|
Indeed, fresh from the needle! |
|
That is so great, it's going to be really powerful! |
|
Is this much faster than |
|
How does this look? library(stars)
# Loading required package: abind
# Loading required package: sf
# Linking to GEOS 3.5.1, GDAL 2.4.0dev-04296c6, PROJ 5.1.0
tif = system.file("tif/L7_ETMs.tif", package = "stars")
x = read_stars(tif)[,1:100,1:100,6] # 100 x 100 area
summary(x[[1]])
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 11.00 32.00 41.00 49.09 59.00 255.00
breaks = c(0, 70, 90, 255)
system.time(s1 <- st_as_sf(cut(x, breaks = breaks)))
# user system elapsed
# 0.090 0.004 0.093
system.time(s2 <- st_as_sf(x, as_points = TRUE, breaks = breaks))
# user system elapsed
# 0.38 0.00 0.38 |
|
Personally I am not a fan of contouring gridded data, it seems an unnecessary tinkering that can sometimes lead to misunderstandings (e.g.: it hides what the real resolution of the data is). |
|
Thanks! I think we need to distinguish between resolution (# of pixels per area or length unit) and support (the size of the element a pixel refers to). Yes, OK to close here. |
|
I'm used to dealing with climate models (so the value is an aggregate over the pixel area) but I see your point, makes a lot of sense. Thanks. |



From #12 I performed some tests on the performance of different polygonizing methods for raster data, both for
starsand forrasterdata.The input files are the same used in the first blog post about
stars, that is the NetCDF file fromftp://ftp.dwd.de/pub/data/gpcc/full_data_daily_V1/full_data_daily_2013.nc.gz.Code:
Result:
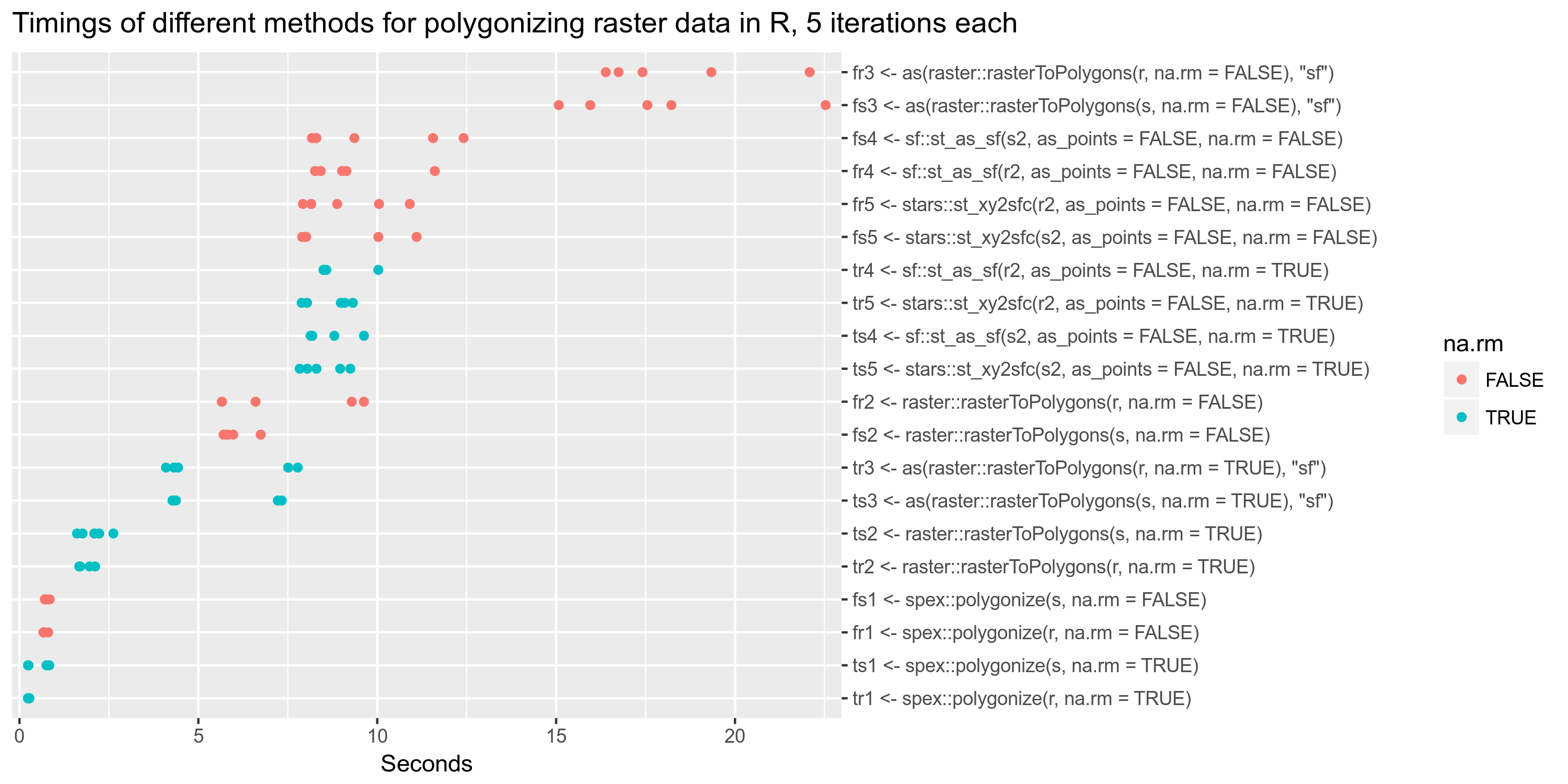
spex::polygonizeis by far the fastest. The methods fromsfandstarsalso are missingna.rmso they take a performance hit on a dataset with many NAs, but in any case they do not outperform the other methods. Note that each different function outputs slightly different data: some outputSpatialPolygonsDataFrame, somesfand somestarsobjects.Overall
spex::polygonizeis not only the fastest but also the most convenient, outputting a clean simple features collection with a different column for each band (timestep).Note that, as it should be, polygonizing a multi-band file (
s,s2) has almost no performance hit over a single-band file (r,r2).The text was updated successfully, but these errors were encountered: