-
Notifications
You must be signed in to change notification settings - Fork 5
Autocycler cluster
The autocycler cluster command groups input contigs into clusters. A cluster is a group of contigs which represent the same genomic sequence. It also decides which of these clusters should be included in the final assembly (QC-pass) and which should not (QC-fail).
For example, in a bacterial genome with a chromosome and a plasmid, each input assembly should ideally contain a contig for the complete chromosome and a contig for the complete plasmid. This step should produce a cluster of chromosome contigs and a cluster of plasmid contigs, both of which should pass QC. Any additional contigs which do not cluster well (e.g. fragmented pieces of the genome from an input assembly that failed to complete) will end up in clusters that fail QC.
autocycler cluster -a autocycler_outThis command takes a directory created by Autocycler compress (named autocycler in the example) which must contain an input_assemblies.gfa file. It will create a subdirectory named clustering which in turn contains a qc_pass directory for the good clusters and a qc_fail directory for the bad clusters.
Usage: autocycler cluster [OPTIONS] --autocycler_dir <AUTOCYCLER_DIR>
Options:
-a, --autocycler_dir <AUTOCYCLER_DIR>
Autocycler directory containing input_assemblies.gfa file (required)
--cutoff <CUTOFF>
cutoff distance threshold for hierarchical clustering [default: 0.2]
--min_assemblies <MIN_ASSEMBLIES>
exclude clusters with fewer than this many assemblies [default: automatic]
--max_contigs <MAX_CONTIGS>
refuse to run if mean contigs per assembly exceeds this value [default: 25]
--manual <MANUAL>
manually define clusters using tree node numbers [default: automatic]
-h, --help
Print help
-V, --version
Print version
- The
--max_contigsoption exists to catch obviously bad input data. If the mean number of contigs per input assemblies exceeds this value (default of 25), Autocycler cluster will refuse to run and display an error message. For example, if you give Autocycler 10 input assemblies with a total of 1000 contigs, that is an average of 100 contigs per assembly, which almost certainly means that they are fragmented or contaminated and thus not appropriate for Autocycler.
There are two reasons that a cluster might fail QC:
-
Present in too few assemblies: the cluster is only found in a small number of assemblies. This threshold is controlled by the
--min_assembliesoption. If not explicitly set,--min_assembliesis set with the following logic: If there is just 1 input assembly (unusual) then set to 1. Otherwise, set to one-quarter of the assembly count (rounded) but no less than 2. - Contained within another cluster: the cluster is contained within another larger cluster which has passed QC. This is to exclude clusters which contain a fragmented piece of a sequence. For example, consider input assemblies where most contain a complete chromosome but some contain the chromosome fragmented into two contigs. This will likely result in three clusters: complete chromosome, chromosome piece 1 and chromosome piece 2. The two chromosome piece clusters will fail QC because their sequences are contained within the larger complete chromosome cluster.
To calculate the pairwise distance between contig A and contig B, Autocycler uses this formula: 1 - (A ∩ B) / A, where A ∩ B is the total length of all unitigs shared by both contigs and A is the total length of all unitigs in the first contig.. This means that the distances are asymmetric: the A-vs-B distance is likely different from the B-vs-A distance. You can see this if you view the clustering/pairwise_distances.phylip file, for example:
3
assembly_01.fasta contig_a (1044289 bp) 0.00000000 0.00052292 0.31332140
assembly_02.fasta contig_b (1044294 bp) 0.00052388 0.00000000 0.31329124
assembly_03.fasta contig_c (713618 bp) 0.00047100 0.00042614 0.00000000
In this case, contig_a-vs-contig_c has a large distance of 0.31, but contig_c-vs-contig_a has a small distance of 0.00047. This is because contig_c is contained within contig_a.
Autocycler uses this asymmetry to see whether a contig is contained within another contig, which is used for QCing clusters (see above). Before running UPGMA clustering, the matrix is made symmetrical by taking the mean distance for each pair.
Clusters are first defined using a threshold on the UPGMA tree, set using the --cutoff option. They are then are iteratively refined: Autocycler tries splitting each cluster to see if the overall clustering score improves (see Clustering metrics for an explanation of the score).
This cluster-splitting refinement can potentially improve the clustering score in two main ways:
- If the genome contains two very similar sequences (e.g. two plasmids which share a lot of sequence), they may initially be grouped into a single cluster. Cluster refinement can divide such a fused cluster into two separate clusters.
- A cluster may contain an outlier contig, e.g. most contigs contain a full chromosomal sequence but one contig only contains 90% of the chromosome. Cluster refinement can exclude the outlier in such a case to produce a tighter cluster.
Subsequent Autocycler steps operate only on QC-pass clusters and ignore QC-fail clusters. You can therefore move cluster directories between the clustering/qc_pass and clustering/qc_fail directories to include/exclude them from the consensus assembly.
Autocycler cluster saves its UPGMA clustering tree in the clustering/clustering.newick file. You can view this tree (e.g. with Taxonium) to better understand the clusters. If you would like to manually specify which clades of the tree should be used to define clusters, their numbers can be given using the --manual option, e.g. --manual 31,38. Hovering over a clade root in Taxonium will display its number.
When examining the clustering, you might find that particular assemblers consistently struggle with a given genome (e.g. miniasm and Raven always produce fragmented assemblies). When this happens, you can go back to the Generating input assemblies and change which assemblers you use (e.g. replace miniasm and Raven with alternative assemblers).
(The toy example is introduced on the Autocycler compress page.)
Running autocycler cluster on the toy example's unitig graph produces the following distance matrix for all input contigs:
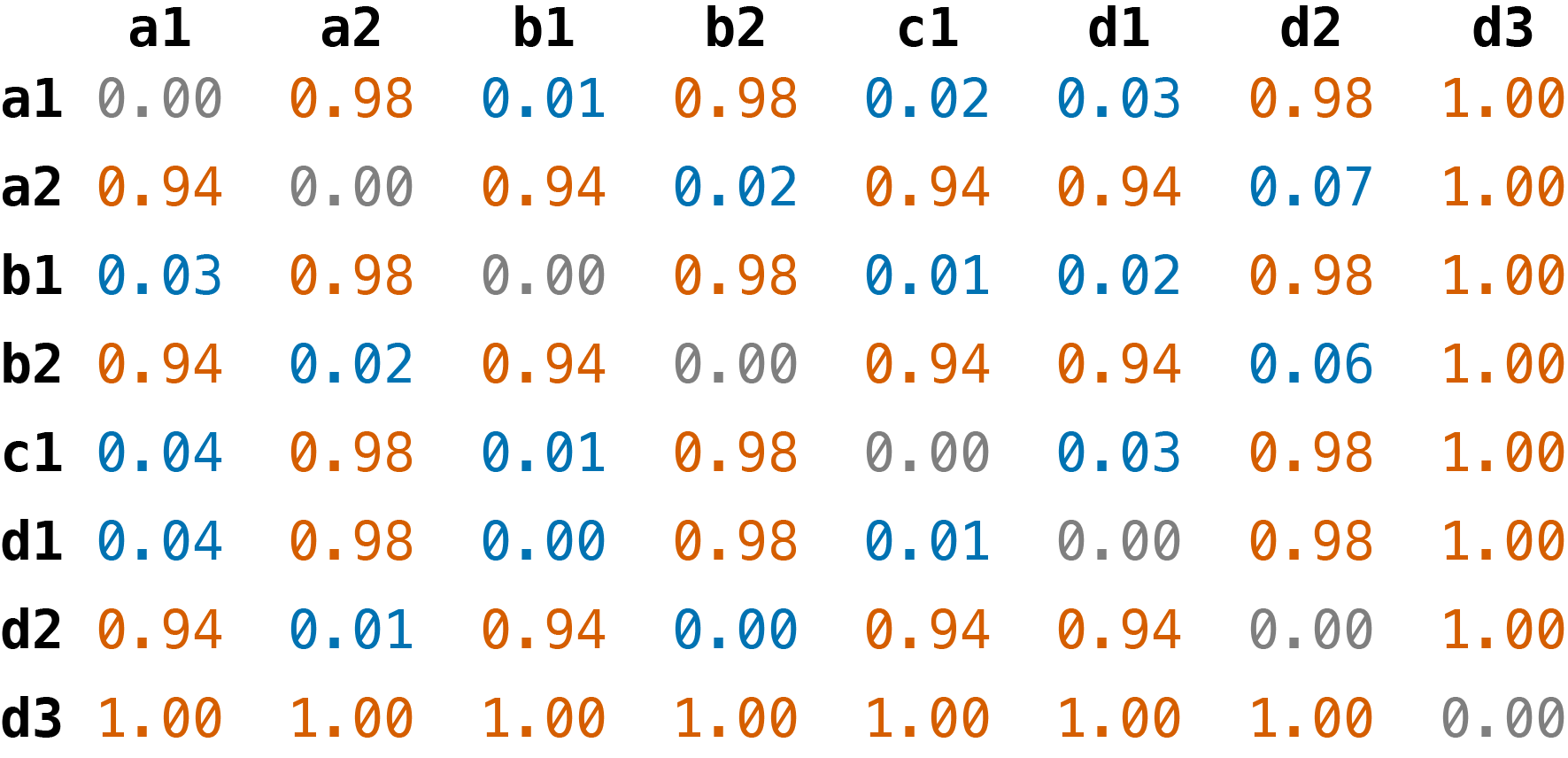
These distances are made by comparing each contig's set of unitigs in the graph, not by comparing sequences directly, making it very fast to compute. Each distance is calculated as 1 - (A ∩ B) / A, where A ∩ B is the total length of all unitigs shared by both contigs and A is the total length of all unitigs in the first contig. Note that this formula creates an asymmetric distance matrix which is useful because it allows us to distinguish when one contig is contained within another. For example, all of d2's unitigs are contained in b2's path but b2 also contains unitigs that are not in d2's path. These means that the d2-vs-b2 distance is 0 while the b2-vs-d2 distance is 0.06.
These distances are then used to perform UPGMA clustering:
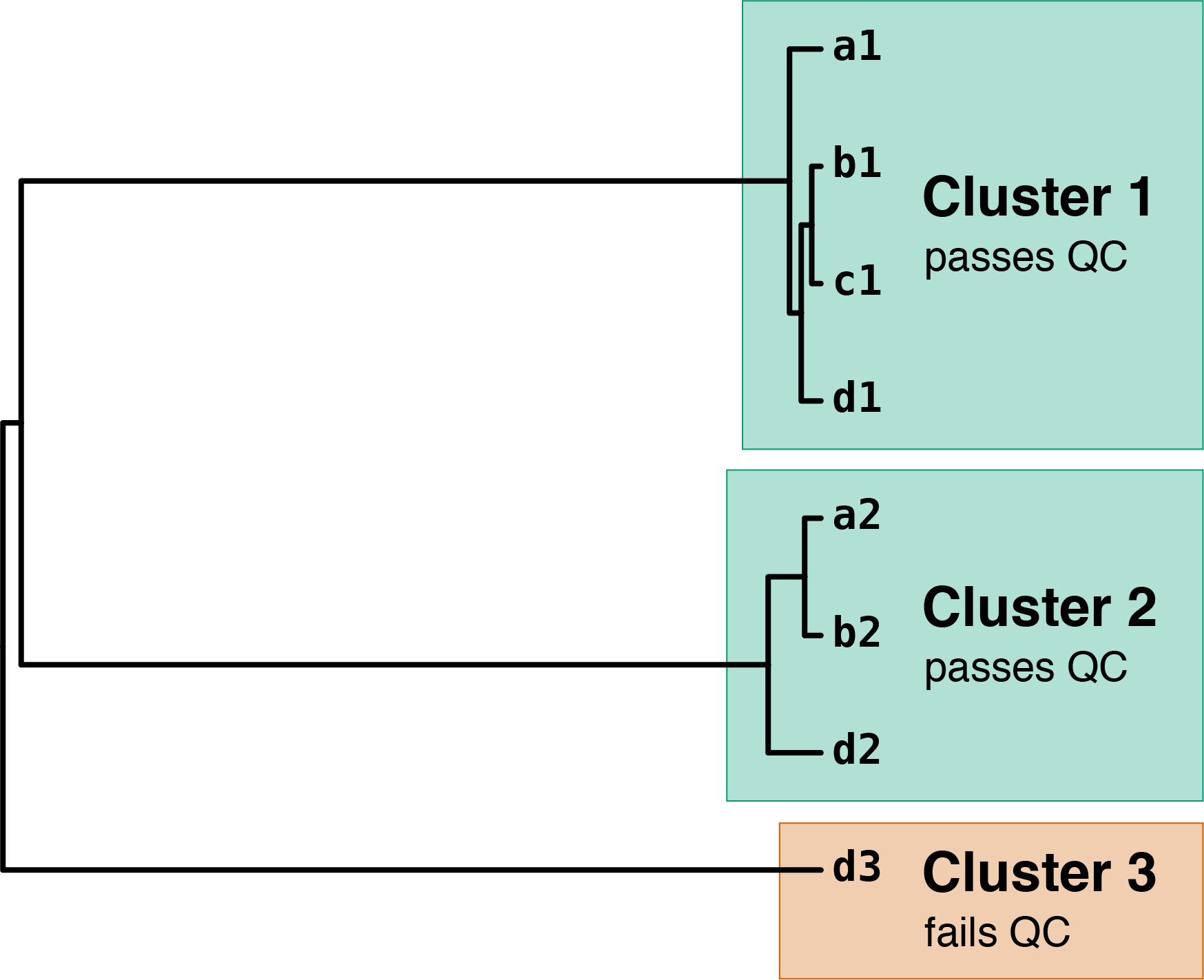
Contigs a1, b1, c1 and d1 are all close in the UPGMA tree, producing cluster 1. Contigs a2, b2 and d2 also form a close group (cluster 2), despite not having a contig from all of the assemblies.
Contig d3 has no close neighbours in the tree, so it forms cluster 3 by itself. Since this cluster only contains a contig from one assembly (below the --min_assemblies threshold), it fails QC and will not be used in subsequent steps.
For each cluster, its unitigs are saved as a separate graph named 1_untrimmed.gfa. Here are the graphs for the two QC-pass clusters in this example:
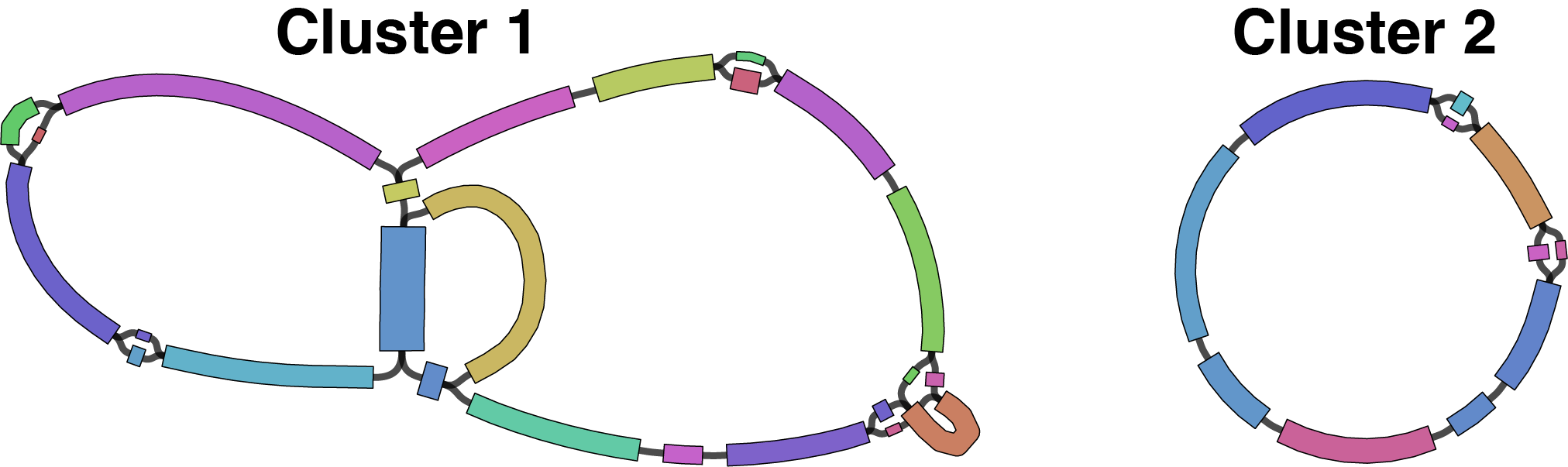
The next two steps in the process (Autocycler trim and Autocycler resolve) operate on a per-cluster basis using these graphs.
- Step 1: Autocycler subsample
- Step 2: Generating input assemblies
- Step 3: Autocycler compress
- Step 4: Autocycler cluster
- Step 5: Autocycler trim
- Step 6: Autocycler resolve
- Step 7: Autocycler combine