-
Notifications
You must be signed in to change notification settings - Fork 28
How circularisation repair works
Most bacterial replicons are circular, which is relevant for Trycycler in two ways: getting a clean circularisation (no gap or overlap) and getting a consistent starting point. This is done as part of the Trycycler reconcile command.
Trycycler attempts to circularise each contig sequence using each of the other sequences as a reference. Specifically, it searches for the start and end of the contig in the other sequences and uses that to determine whether the contig is already circular, needs sequence added or needs sequence removed.
In the following examples, sequence A is the one we are trying to circularise and sequence B is the other reference sequence.
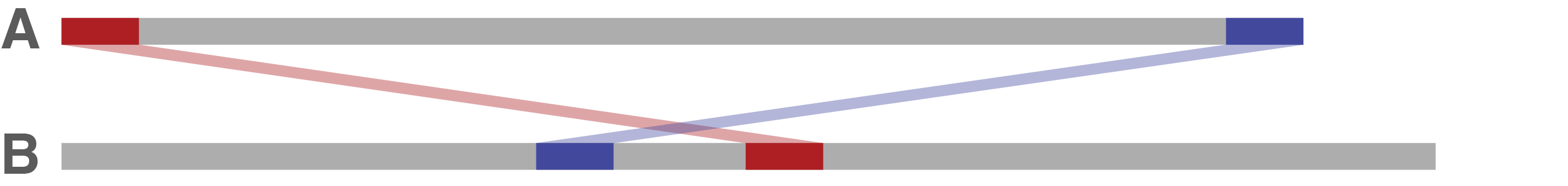
Ideally, sequence A's end is immediately followed by sequence A's start in sequence B. If this is the case, that means sequence A is already circular and there's nothing more to do.
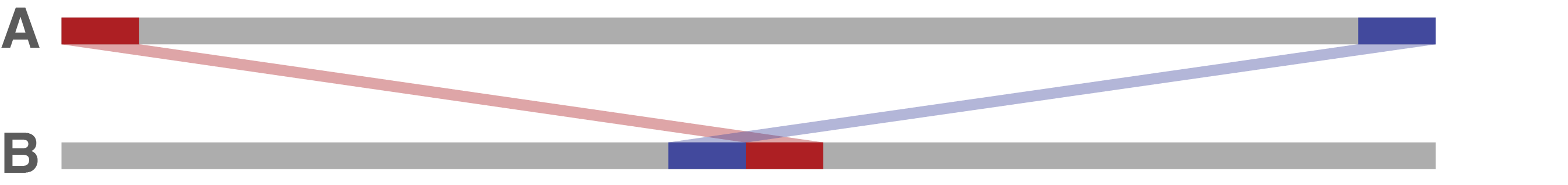
It may be that sequence A's end and start are both found in sequence B, but with a gap in between. This implies that sequence A is missing some sequence in its circularisation. Trycycler will fill in this gap using the sequence between the hits in sequence B.
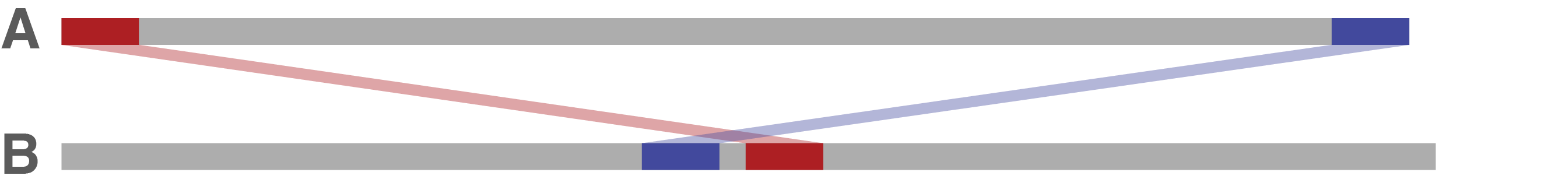
If sequence A's end and start overlap in sequence B, that implies that sequence A has too much sequence – i.e. some sequence is duplicated at its start/end. In this case, Trycycler will trim sequence A's end to give it a clean circularisation.
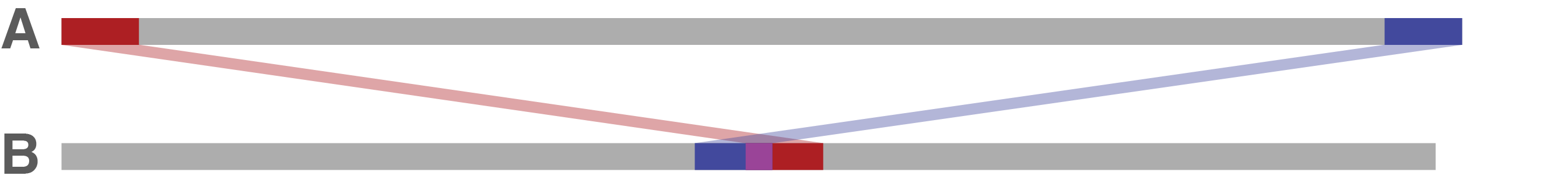
If there is too much gap between sequence A's end and start in sequence B, that implies that sequence A is missing a lot of sequence. Trycycler will fail to circularise A in this case. It probably makes sense to exclude sequence A and try running Trycycler reconcile again.

Conversely, sequence A's start might come well before sequence A's end in sequence B. This implies that sequence A has quite a lot of overlap. Trycycler may be able to resolve this by trimming the start/end of sequence A, but it might not. If this happens, you can try to manually trim sequence A and then try running Trycycler reconcile again. Or else you can simply exclude sequence A.
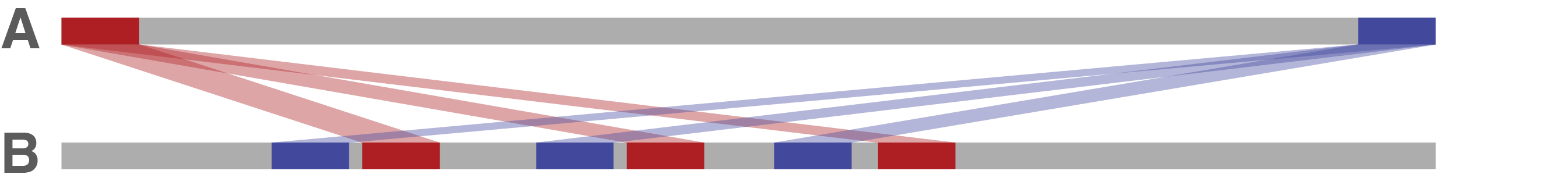
If sequence A's start and end are found in multiple places in sequence B, this will also cause Trycycler to fail circularisation. This suggests that sequence A begins/end in a repeat sequence – not necessarily a problem with the assembly but it does make circularisation difficult. Trycycler can sometimes resolve this (by trimming sequence A's start/end) but not always. In cases where multiple hits cause a circularisation failure, simply excluding sequence A is probably in order.

If sequence A start or end is not found in sequence B, that will also cause a failure to circularise. This suggests that either sequence A contains spurious sequence or sequence B contains missing sequence. When this causes a circularisation failure, it's best to exclude sequence A.

If sequence A and sequence B have the same start/end, then there is no information for fixing A's circularisation. This sometimes happens with two input assemblies from the same assembler. It's usually not a problem, as sequence A's circularisation be repaired using one of the other sequences instead.
Trycycler will conduct all pairwise circularisation. For example, if you have 4 input assemblies (A, B, C and D), Trycycler will attempt to circularise sequence A using sequences B, C and D. It will attempt to circularise sequence B using sequences A, C and D. And so on.
This means there can be multiple ways to circularise a sequence. For example, sequence A might be circularised in three ways: 20 bp added from B, 21 bp added from C and 19 bp added from D. To choose which is the best option, Trycycler aligns the reads to the circularisation junction (this is why reads must be given as a command line parameter to Trycycler reconcile). Whichever circularisation option results in the highest total alignment score is chosen as the final one.
A circular sequence can potentially start at any point on either strand and still be a valid assembly. However, when reconciling multiple alternative contigs, it is necessary to make all sequences consistent with each other – i.e. start at the same point and on the same strand.

By convention, Trycycler will try to start the contigs at a replication initiator protein gene sequence like dnaA. To be a suitable starting point, the starting sequence must be in each of the contigs and only occur once in each contig.
If a replication initiator protein gene sequence can't be found, Trycycler will randomly select a subsequence which is present in each of the contigs only once and use that as the starting sequence.
- Home
- Software requirements
- Installation
-
How to run Trycycler
- Quick start
- Step 1: Generating assemblies
- Step 2: Clustering contigs
- Step 3: Reconciling contigs
- Step 4: Multiple sequence alignment
- Step 5: Partitioning reads
- Step 6: Generating a consensus
- Step 7: Polishing after Trycycler
- Illustrated pipeline overview
- Demo datasets
- Implementation details
- FAQ and miscellaneous tips
- Other pages
- Guide to bacterial genome assembly (choose your own adventure)
- Accuracy vs depth