-
Notifications
You must be signed in to change notification settings - Fork 28
Mediocre dataset analysis
To begin, I ran Trycycler cluster on the assemblies:
trycycler cluster --reads reads.fastq.gz --assemblies assemblies/*.fasta --out_dir trycyclerWhich produced this output.
There were nine clusters, but everything but the first two only contain a single contig. Here's the tree:
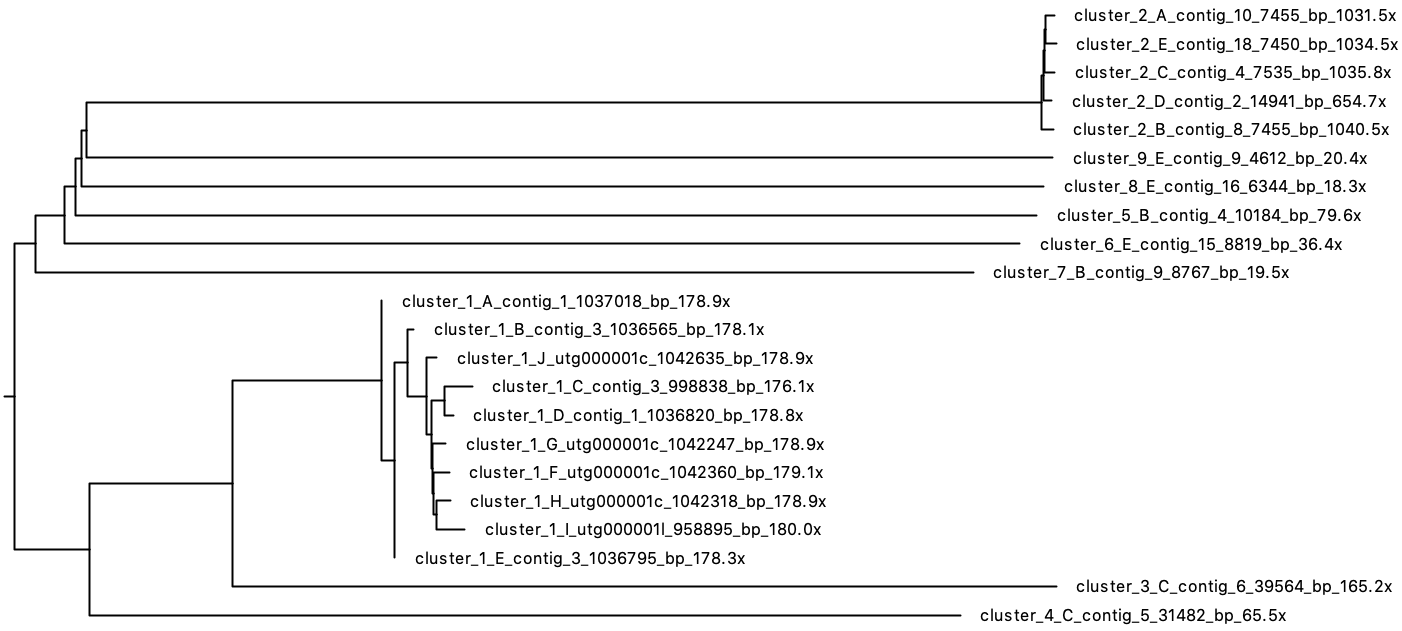
I therefore conclude that cluster 1 is the chromosome and cluster 2 is the plasmid. All other clusters look like junk so I get rid of them:
mv trycycler/cluster_003 trycycler/bad_cluster_003
mv trycycler/cluster_004 trycycler/bad_cluster_004
mv trycycler/cluster_005 trycycler/bad_cluster_005
mv trycycler/cluster_006 trycycler/bad_cluster_006
mv trycycler/cluster_007 trycycler/bad_cluster_007
mv trycycler/cluster_008 trycycler/bad_cluster_008
mv trycycler/cluster_009 trycycler/bad_cluster_009Prefixing the clusters with bad_ will allow me to later glob for all good clusters with trycycler/cluster_*. Alternatively, you could just delete the bad cluster directories.
I then ran Trycycler reconcile on the first (chromosomal) cluster:
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_001Which produced this output. One of the sequences (F_utg000001c) failed to circularise due to multiple start/end hits, so I removed it and ran Trycycler reconcile again:
mv trycycler/cluster_001/1_contigs/F_utg000001c.fasta trycycler/cluster_001/1_contigs/F_utg000001c.fasta.bad
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_001Which produced this output. Now a different sequence (I_utg000001l) failed to circularise, this time due to a too-large start/end gap (consistent with its shorter length). I removed it and ran Trycycler reconcile again:
mv trycycler/cluster_001/1_contigs/I_utg000001l.fasta trycycler/cluster_001/1_contigs/I_utg000001l.fasta.bad
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_001Which produced this output. Nearly there! The circularisation completed, but one of the sequences (C_contig_3) looked terrible in the final alignment check. So I removed it and ran Trycycler reconcile again:
mv trycycler/cluster_001/1_contigs/C_contig_3.fasta trycycler/cluster_001/1_contigs/C_contig_3.fasta.bad
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_001Which produced this output. Finally, cluster 1 is reconciled!
I then ran Trycycler reconcile on the second (plasmid) cluster:
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_002Which produced this output. We encountered the same problem as in the Good dataset analysis, where there was a doubled-version of the plasmid in one of the contigs. I manually repaired that contig and ran Trycycler reconcile again:
trycycler reconcile --reads reads.fastq.gz --cluster_dir trycycler/cluster_002Which produced this output. Everything looks good this time!
Since these final three steps usually don't require human intervention, I'll run them all at once here. I'll also use Bash loops to run the cluster-specific commands on each cluster. While this may look a bit clunky, it's nice and reusable as it will work for Trycycler genomes with any number of clusters:
for c in trycycler/cluster_*; do
trycycler msa --cluster_dir "$c"
done
trycycler partition --reads reads.fastq.gz --cluster_dirs trycycler/cluster_*
for c in trycycler/cluster_*; do
trycycler consensus --cluster_dir "$c"
done
cat trycycler/cluster_*/7_final_consensus.fasta > assembly.fastaThere are the outputs: cluster 1 msa, cluster 2 msa, partition, cluster 1 consensus and cluster 2 consensus.
This genome had a few snags, but we were still able to get a nice assembly. The final sequences contain no significant errors: just some small-scale mistakes and wrong homopolymer lengths.
- Home
- Software requirements
- Installation
-
How to run Trycycler
- Quick start
- Step 1: Generating assemblies
- Step 2: Clustering contigs
- Step 3: Reconciling contigs
- Step 4: Multiple sequence alignment
- Step 5: Partitioning reads
- Step 6: Generating a consensus
- Step 7: Polishing after Trycycler
- Illustrated pipeline overview
- Demo datasets
- Implementation details
- FAQ and miscellaneous tips
- Other pages
- Guide to bacterial genome assembly (choose your own adventure)
- Accuracy vs depth