-
Notifications
You must be signed in to change notification settings - Fork 20
fitness_landscape_timing
Daniel Falster edited this page Jun 28, 2024
·
2 revisions
library(tree)p <- ebt_base_parameters()p$add_strategy(new(Strategy, list(lma=0.0648406)))p$add_strategy(new(Strategy, list(lma=0.1977910)))p$seed_rain <- c(1.1, 1.1) # Starting rain.Work out what the equilibrium seed rain is. This takes a while! If using the second set of seed_rain values this should converge quickly once the cohort schedule is worked out.
res <- equilibrium_seed_rain(p)## 1: Splitting {9,23} times (141,141)
## 2: Splitting {1,3} times (150,164)
## *** 1: {1.1,1.1} -> {7.738,29.13} (delta = {6.638,28.03})
## 1: Splitting {15,25} times (141,141)
## 2: Splitting {4,28} times (156,166)
## 3: Splitting {1,12} times (160,194)
## 4: Splitting {0,6} times (161,206)
## *** 2: {7.738,29.13} -> {7.976,23.42} (delta = {0.2382,-5.71})
## *** 3: {7.976,23.42} -> {8,23.24} (delta = {0.02382,-0.1779})
## *** 4: {8,23.24} -> {8.001,23.23} (delta = {0.0008665,-0.01203})
## *** 5: {8.001,23.23} -> {8.001,23.23} (delta = {6.738e-05,-0.0004422})
## *** 6: {8.001,23.23} -> {8.001,23.23} (delta = {1.955e-06,-3.372e-05})
## *** 7: {8.001,23.23} -> {8.001,23.23} (delta = {1.191e-07,-1.521e-06})
## Reached target accuracy (delta 1.52064e-06 < 1.00000e-05 eps)
Set the seed rain back into the parameters
p$seed_rain <- res$seed_rain[,"out"]schedule <- res$scheduleResident lma values:
lma_res <- sapply(seq_len(p$size), function(i) p[[i]]$parameters$lma)OK, good to reasonable accuracy; the seed rain values should be close enough to 1.
cmp <- fitness_landscape("lma", lma_res, p, schedule)cmp # This should be about zero, plus or minus 1e-5 or so.## [1] -2.467e-06 -1.668e-03
# Mutant LMA values, in increasing numbers, to test how the time# requirements scale with the number of strategies. Should be# sublinear.seq_log <- function(from, to, length.out) {
exp(seq(log(from), log(to), length.out=length.out))
}lma_2 <- seq_log(0.03, 0.8, 2)lma_4 <- seq_log(0.03, 0.8, 4)lma_8 <- seq_log(0.03, 0.8, 8)lma_16 <- seq_log(0.03, 0.8, 16)lma_32 <- seq_log(0.03, 0.8, 32)lma_64 <- seq_log(0.03, 0.8, 64)(t_2 <- system.time(w_2 <- fitness_landscape("lma", lma_2, p, schedule)))## user system elapsed
## 17.196 0.033 17.310
(t_4 <- system.time(w_4 <- fitness_landscape("lma", lma_4, p, schedule)))## user system elapsed
## 25.369 0.052 25.545
(t_8 <- system.time(w_8 <- fitness_landscape("lma", lma_8, p, schedule)))## user system elapsed
## 40.851 0.078 41.111
(t_16 <- system.time(w_16 <- fitness_landscape("lma", lma_16, p, schedule)))## user system elapsed
## 75.004 0.198 76.102
(t_32 <- system.time(w_32 <- fitness_landscape("lma", lma_32, p, schedule)))## user system elapsed
## 161.520 0.434 164.307
(t_64 <- system.time(w_64 <- fitness_landscape("lma", lma_64, p, schedule)))## user system elapsed
## 311.256 0.874 317.624
tt <- c(t_2[["elapsed"]],
t_4[["elapsed"]],
t_8[["elapsed"]],
t_16[["elapsed"]],
t_32[["elapsed"]],
t_64[["elapsed"]])n <- 2^seq_along(tt)This is largely OK, but the increased per-mutant time is concerning (might be due to running other things at the same time though)
plot(n, tt / n, log="x")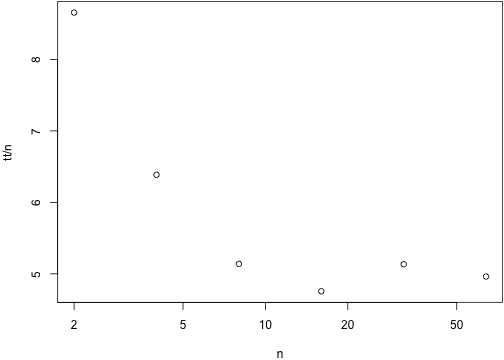
plot(w_64 ~ lma_64, type="l", log="x")abline(v=lma_res, lty=3, col="grey")
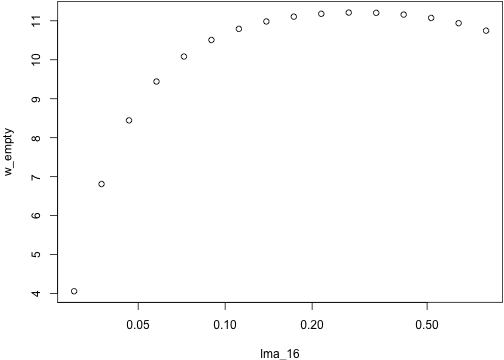
# Fitness of the mutants in an empty landscape:w_empty <- fitness_landscape_empty("lma", lma_16, p, schedule)plot(w_empty ~ lma_16, log="x")